Victor M. Campello
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Intrapartum Ultrasound Image Segmentation of Pubic Symphysis and Fetal Head Using Dual Student-Teacher Framework with CNN-ViT Collaborative Learning
Sep 11, 2024Abstract:The segmentation of the pubic symphysis and fetal head (PSFH) constitutes a pivotal step in monitoring labor progression and identifying potential delivery complications. Despite the advances in deep learning, the lack of annotated medical images hinders the training of segmentation. Traditional semi-supervised learning approaches primarily utilize a unified network model based on Convolutional Neural Networks (CNNs) and apply consistency regularization to mitigate the reliance on extensive annotated data. However, these methods often fall short in capturing the discriminative features of unlabeled data and in delineating the long-range dependencies inherent in the ambiguous boundaries of PSFH within ultrasound images. To address these limitations, we introduce a novel framework, the Dual-Student and Teacher Combining CNN and Transformer (DSTCT), which synergistically integrates the capabilities of CNNs and Transformers. Our framework comprises a Vision Transformer (ViT) as the teacher and two student mod ls one ViT and one CNN. This dual-student setup enables mutual supervision through the generation of both hard and soft pseudo-labels, with the consistency in their predictions being refined by minimizing the classifier determinacy discrepancy. The teacher model further reinforces learning within this architecture through the imposition of consistency regularization constraints. To augment the generalization abilities of our approach, we employ a blend of data and model perturbation techniques. Comprehensive evaluations on the benchmark dataset of the PSFH Segmentation Grand Challenge at MICCAI 2023 demonstrate our DSTCT framework outperformed ten contemporary semi-supervised segmentation methods. Code available at https://github.com/jjm1589/DSTCT.
Cardiac Segmentation on Late Gadolinium Enhancement MRI: A Benchmark Study from Multi-Sequence Cardiac MR Segmentation Challenge
Jun 22, 2020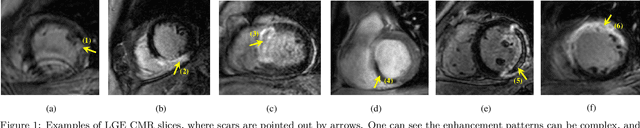
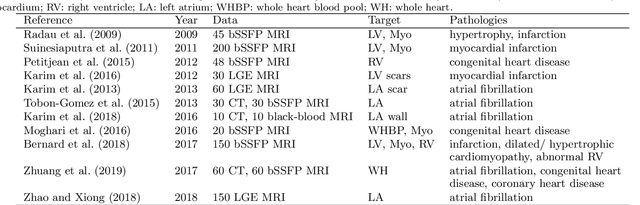
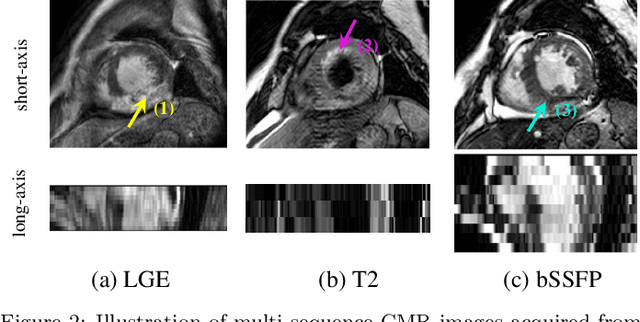
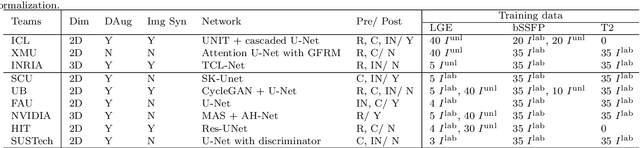
Abstract:Accurate computing, analysis and modeling of the ventricles and myocardium from medical images are important, especially in the diagnosis and treatment management for patients suffering from myocardial infarction (MI). Late gadolinium enhancement (LGE) cardiac magnetic resonance (CMR) provides an important protocol to visualize MI. However, automated segmentation of LGE CMR is still challenging, due to the indistinguishable boundaries, heterogeneous intensity distribution and complex enhancement patterns of pathological myocardium from LGE CMR. Furthermore, compared with the other sequences LGE CMR images with gold standard labels are particularly limited, which represents another obstacle for developing novel algorithms for automatic segmentation of LGE CMR. This paper presents the selective results from the Multi-Sequence Cardiac MR (MS-CMR) Segmentation challenge, in conjunction with MICCAI 2019. The challenge offered a data set of paired MS-CMR images, including auxiliary CMR sequences as well as LGE CMR, from 45 patients who underwent cardiomyopathy. It was aimed to develop new algorithms, as well as benchmark existing ones for LGE CMR segmentation and compare them objectively. In addition, the paired MS-CMR images could enable algorithms to combine the complementary information from the other sequences for the segmentation of LGE CMR. Nine representative works were selected for evaluation and comparisons, among which three methods are unsupervised methods and the other six are supervised. The results showed that the average performance of the nine methods was comparable to the inter-observer variations. The success of these methods was mainly attributed to the inclusion of the auxiliary sequences from the MS-CMR images, which provide important label information for the training of deep neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge