Sulaiman Vesal
Fine-tuning Small Language Models as Efficient Enterprise Search Relevance Labelers
Jan 06, 2026Abstract:In enterprise search, building high-quality datasets at scale remains a central challenge due to the difficulty of acquiring labeled data. To resolve this challenge, we propose an efficient approach to fine-tune small language models (SLMs) for accurate relevance labeling, enabling high-throughput, domain-specific labeling comparable or even better in quality to that of state-of-the-art large language models (LLMs). To overcome the lack of high-quality and accessible datasets in the enterprise domain, our method leverages on synthetic data generation. Specifically, we employ an LLM to synthesize realistic enterprise queries from a seed document, apply BM25 to retrieve hard negatives, and use a teacher LLM to assign relevance scores. The resulting dataset is then distilled into an SLM, producing a compact relevance labeler. We evaluate our approach on a high-quality benchmark consisting of 923 enterprise query-document pairs annotated by trained human annotators, and show that the distilled SLM achieves agreement with human judgments on par with or better than the teacher LLM. Furthermore, our fine-tuned labeler substantially improves throughput, achieving 17 times increase while also being 19 times more cost-effective. This approach enables scalable and cost-effective relevance labeling for enterprise-scale retrieval applications, supporting rapid offline evaluation and iteration in real-world settings.
Learning ECG Representations via Poly-Window Contrastive Learning
Aug 21, 2025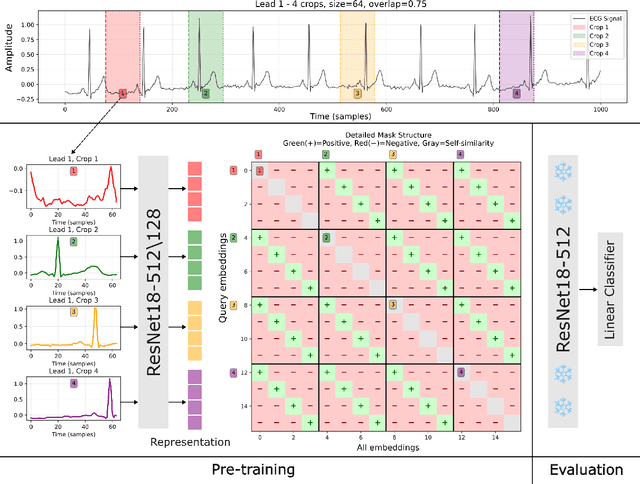
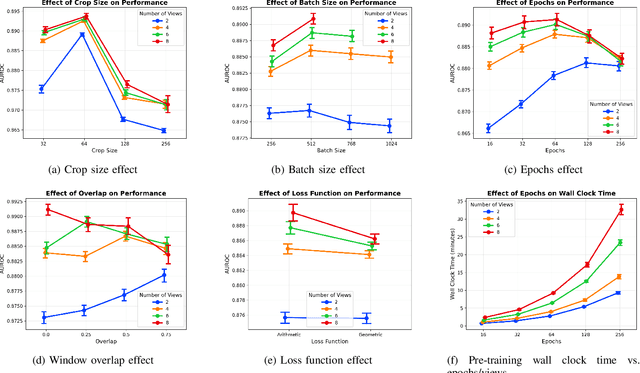
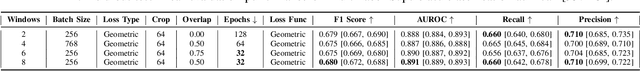

Abstract:Electrocardiogram (ECG) analysis is foundational for cardiovascular disease diagnosis, yet the performance of deep learning models is often constrained by limited access to annotated data. Self-supervised contrastive learning has emerged as a powerful approach for learning robust ECG representations from unlabeled signals. However, most existing methods generate only pairwise augmented views and fail to leverage the rich temporal structure of ECG recordings. In this work, we present a poly-window contrastive learning framework. We extract multiple temporal windows from each ECG instance to construct positive pairs and maximize their agreement via statistics. Inspired by the principle of slow feature analysis, our approach explicitly encourages the model to learn temporally invariant and physiologically meaningful features that persist across time. We validate our approach through extensive experiments and ablation studies on the PTB-XL dataset. Our results demonstrate that poly-window contrastive learning consistently outperforms conventional two-view methods in multi-label superclass classification, achieving higher AUROC (0.891 vs. 0.888) and F1 scores (0.680 vs. 0.679) while requiring up to four times fewer pre-training epochs (32 vs. 128) and 14.8% in total wall clock pre-training time reduction. Despite processing multiple windows per sample, we achieve a significant reduction in the number of training epochs and total computation time, making our method practical for training foundational models. Through extensive ablations, we identify optimal design choices and demonstrate robustness across various hyperparameters. These findings establish poly-window contrastive learning as a highly efficient and scalable paradigm for automated ECG analysis and provide a promising general framework for self-supervised representation learning in biomedical time-series data.
Registration-Enhanced Segmentation Method for Prostate Cancer in Ultrasound Images
Feb 02, 2025



Abstract:Prostate cancer is a major cause of cancer-related deaths in men, where early detection greatly improves survival rates. Although MRI-TRUS fusion biopsy offers superior accuracy by combining MRI's detailed visualization with TRUS's real-time guidance, it is a complex and time-intensive procedure that relies heavily on manual annotations, leading to potential errors. To address these challenges, we propose a fully automatic MRI-TRUS fusion-based segmentation method that identifies prostate tumors directly in TRUS images without requiring manual annotations. Unlike traditional multimodal fusion approaches that rely on naive data concatenation, our method integrates a registration-segmentation framework to align and leverage spatial information between MRI and TRUS modalities. This alignment enhances segmentation accuracy and reduces reliance on manual effort. Our approach was validated on a dataset of 1,747 patients from Stanford Hospital, achieving an average Dice coefficient of 0.212, outperforming TRUS-only (0.117) and naive MRI-TRUS fusion (0.132) methods, with significant improvements (p $<$ 0.01). This framework demonstrates the potential for reducing the complexity of prostate cancer diagnosis and provides a flexible architecture applicable to other multimodal medical imaging tasks.
Mask Enhanced Deeply Supervised Prostate Cancer Detection on B-mode Micro-Ultrasound
Dec 14, 2024



Abstract:Prostate cancer is a leading cause of cancer-related deaths among men. The recent development of high frequency, micro-ultrasound imaging offers improved resolution compared to conventional ultrasound and potentially a better ability to differentiate clinically significant cancer from normal tissue. However, the features of prostate cancer remain subtle, with ambiguous borders with normal tissue and large variations in appearance, making it challenging for both machine learning and humans to localize it on micro-ultrasound images. We propose a novel Mask Enhanced Deeply-supervised Micro-US network, termed MedMusNet, to automatically and more accurately segment prostate cancer to be used as potential targets for biopsy procedures. MedMusNet leverages predicted masks of prostate cancer to enforce the learned features layer-wisely within the network, reducing the influence of noise and improving overall consistency across frames. MedMusNet successfully detected 76% of clinically significant cancer with a Dice Similarity Coefficient of 0.365, significantly outperforming the baseline Swin-M2F in specificity and accuracy (Wilcoxon test, Bonferroni correction, p-value<0.05). While the lesion-level and patient-level analyses showed improved performance compared to human experts and different baseline, the improvements did not reach statistical significance, likely on account of the small cohort. We have presented a novel approach to automatically detect and segment clinically significant prostate cancer on B-mode micro-ultrasound images. Our MedMusNet model outperformed other models, surpassing even human experts. These preliminary results suggest the potential for aiding urologists in prostate cancer diagnosis via biopsy and treatment decision-making.
Sound event detection based on auxiliary decoder and maximum probability aggregation for DCASE Challenge 2024 Task 4
Jun 17, 2024Abstract:In this report, we propose three novel methods for developing a sound event detection (SED) model for the DCASE 2024 Challenge Task 4. First, we propose an auxiliary decoder attached to the final convolutional block to improve feature extraction capabilities while reducing dependency on embeddings from pre-trained large models. The proposed auxiliary decoder operates independently from the main decoder, enhancing performance of the convolutional block during the initial training stages by assigning a different weight strategy between main and auxiliary decoder losses. Next, to address the time interval issue between the DESED and MAESTRO datasets, we propose maximum probability aggregation (MPA) during the training step. The proposed MPA method enables the model's output to be aligned with soft labels of 1 s in the MAESTRO dataset. Finally, we propose a multi-channel input feature that employs various versions of logmel and MFCC features to generate time-frequency pattern. The experimental results demonstrate the efficacy of these proposed methods in a view of improving SED performance by achieving a balanced enhancement across different datasets and label types. Ultimately, this approach presents a significant step forward in developing more robust and flexible SED models
ProsDectNet: Bridging the Gap in Prostate Cancer Detection via Transrectal B-mode Ultrasound Imaging
Dec 08, 2023



Abstract:Interpreting traditional B-mode ultrasound images can be challenging due to image artifacts (e.g., shadowing, speckle), leading to low sensitivity and limited diagnostic accuracy. While Magnetic Resonance Imaging (MRI) has been proposed as a solution, it is expensive and not widely available. Furthermore, most biopsies are guided by Transrectal Ultrasound (TRUS) alone and can miss up to 52% cancers, highlighting the need for improved targeting. To address this issue, we propose ProsDectNet, a multi-task deep learning approach that localizes prostate cancer on B-mode ultrasound. Our model is pre-trained using radiologist-labeled data and fine-tuned using biopsy-confirmed labels. ProsDectNet includes a lesion detection and patch classification head, with uncertainty minimization using entropy to improve model performance and reduce false positive predictions. We trained and validated ProsDectNet using a cohort of 289 patients who underwent MRI-TRUS fusion targeted biopsy. We then tested our approach on a group of 41 patients and found that ProsDectNet outperformed the average expert clinician in detecting prostate cancer on B-mode ultrasound images, achieving a patient-level ROC-AUC of 82%, a sensitivity of 74%, and a specificity of 67%. Our results demonstrate that ProsDectNet has the potential to be used as a computer-aided diagnosis system to improve targeted biopsy and treatment planning.
Domain Generalization for Prostate Segmentation in Transrectal Ultrasound Images: A Multi-center Study
Sep 05, 2022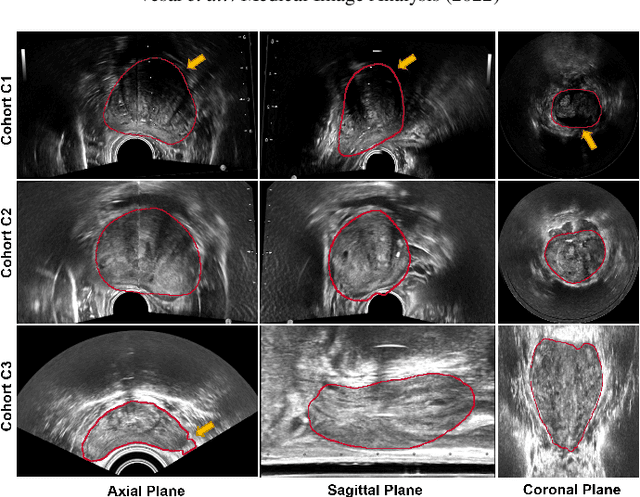
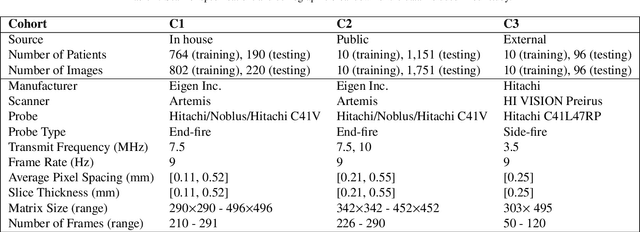
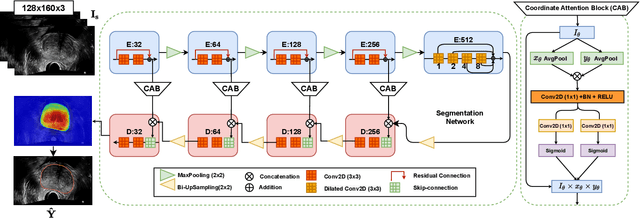
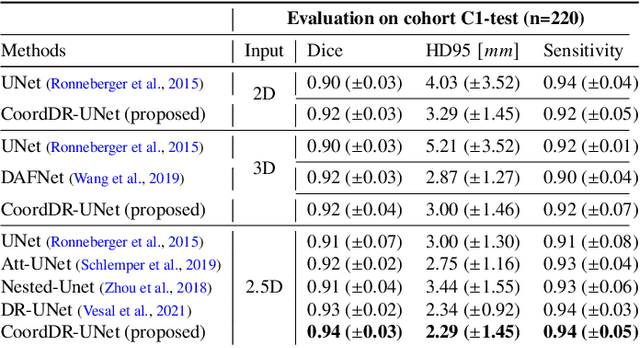
Abstract:Prostate biopsy and image-guided treatment procedures are often performed under the guidance of ultrasound fused with magnetic resonance images (MRI). Accurate image fusion relies on accurate segmentation of the prostate on ultrasound images. Yet, the reduced signal-to-noise ratio and artifacts (e.g., speckle and shadowing) in ultrasound images limit the performance of automated prostate segmentation techniques and generalizing these methods to new image domains is inherently difficult. In this study, we address these challenges by introducing a novel 2.5D deep neural network for prostate segmentation on ultrasound images. Our approach addresses the limitations of transfer learning and finetuning methods (i.e., drop in performance on the original training data when the model weights are updated) by combining a supervised domain adaptation technique and a knowledge distillation loss. The knowledge distillation loss allows the preservation of previously learned knowledge and reduces the performance drop after model finetuning on new datasets. Furthermore, our approach relies on an attention module that considers model feature positioning information to improve the segmentation accuracy. We trained our model on 764 subjects from one institution and finetuned our model using only ten subjects from subsequent institutions. We analyzed the performance of our method on three large datasets encompassing 2067 subjects from three different institutions. Our method achieved an average Dice Similarity Coefficient (Dice) of $94.0\pm0.03$ and Hausdorff Distance (HD95) of 2.28 $mm$ in an independent set of subjects from the first institution. Moreover, our model generalized well in the studies from the other two institutions (Dice: $91.0\pm0.03$; HD95: 3.7$mm$ and Dice: $82.0\pm0.03$; HD95: 7.1 $mm$).
ConFUDA: Contrastive Fewshot Unsupervised Domain Adaptation for Medical Image Segmentation
Jun 08, 2022



Abstract:Unsupervised domain adaptation (UDA) aims to transfer knowledge learned from a labeled source domain to an unlabeled target domain. Contrastive learning (CL) in the context of UDA can help to better separate classes in feature space. However, in image segmentation, the large memory footprint due to the computation of the pixel-wise contrastive loss makes it prohibitive to use. Furthermore, labeled target data is not easily available in medical imaging, and obtaining new samples is not economical. As a result, in this work, we tackle a more challenging UDA task when there are only a few (fewshot) or a single (oneshot) image available from the target domain. We apply a style transfer module to mitigate the scarcity of target samples. Then, to align the source and target features and tackle the memory issue of the traditional contrastive loss, we propose the centroid-based contrastive learning (CCL) and a centroid norm regularizer (CNR) to optimize the contrastive pairs in both direction and magnitude. In addition, we propose multi-partition centroid contrastive learning (MPCCL) to further reduce the variance in the target features. Fewshot evaluation on MS-CMRSeg dataset demonstrates that ConFUDA improves the segmentation performance by 0.34 of the Dice score on the target domain compared with the baseline, and 0.31 Dice score improvement in a more rigorous oneshot setting.
Few-shot Unsupervised Domain Adaptation for Multi-modal Cardiac Image Segmentation
Jan 28, 2022


Abstract:Unsupervised domain adaptation (UDA) methods intend to reduce the gap between source and target domains by using unlabeled target domain and labeled source domain data, however, in the medical domain, target domain data may not always be easily available, and acquiring new samples is generally time-consuming. This restricts the development of UDA methods for new domains. In this paper, we explore the potential of UDA in a more challenging while realistic scenario where only one unlabeled target patient sample is available. We call it Few-shot Unsupervised Domain adaptation (FUDA). We first generate target-style images from source images and explore diverse target styles from a single target patient with Random Adaptive Instance Normalization (RAIN). Then, a segmentation network is trained in a supervised manner with the generated target images. Our experiments demonstrate that FUDA improves the segmentation performance by 0.33 of Dice score on the target domain compared with the baseline, and it also gives 0.28 of Dice score improvement in a more rigorous one-shot setting. Our code is available at \url{https://github.com/MingxuanGu/Few-shot-UDA}.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021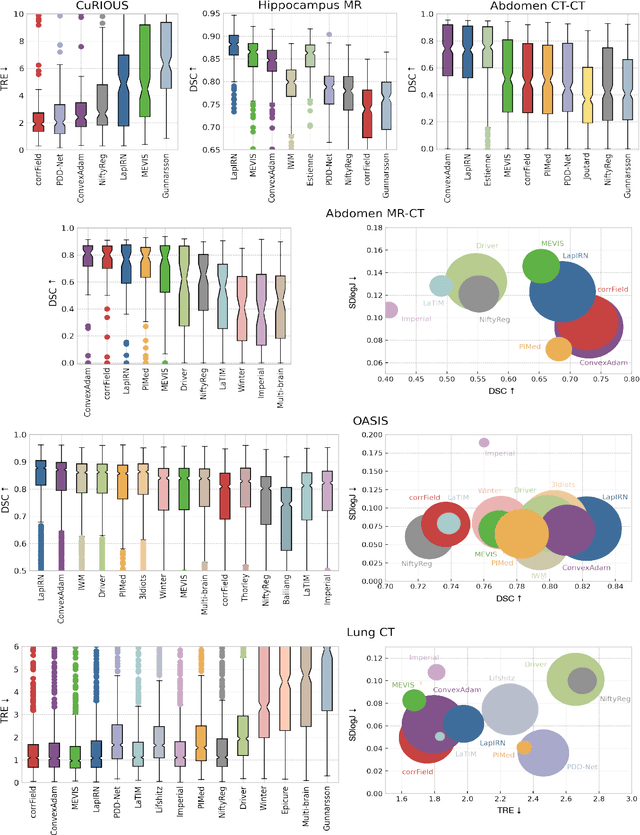
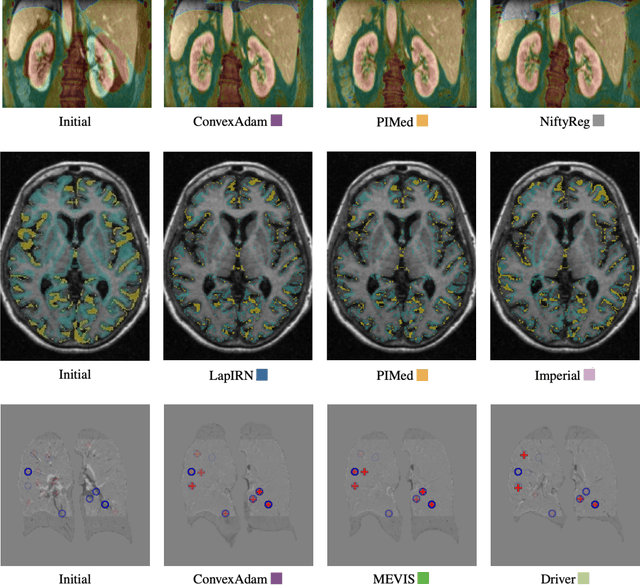
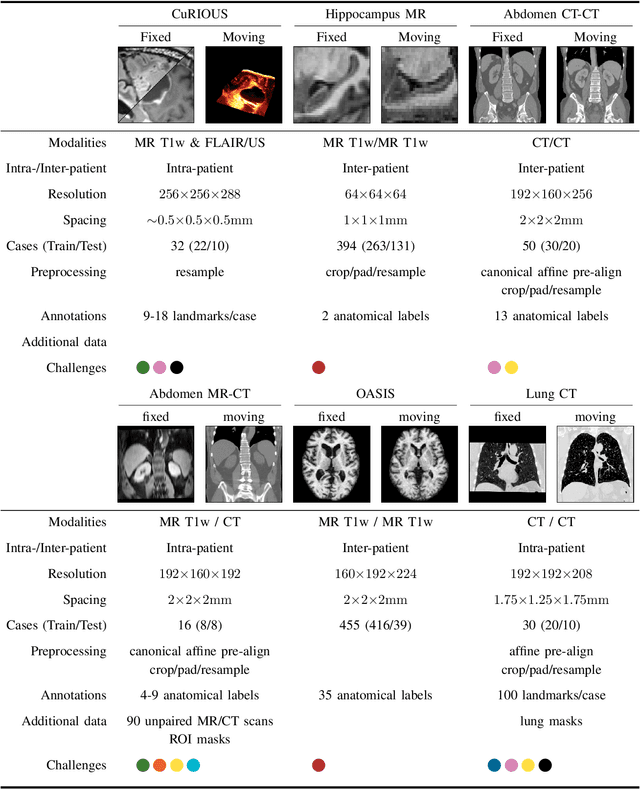
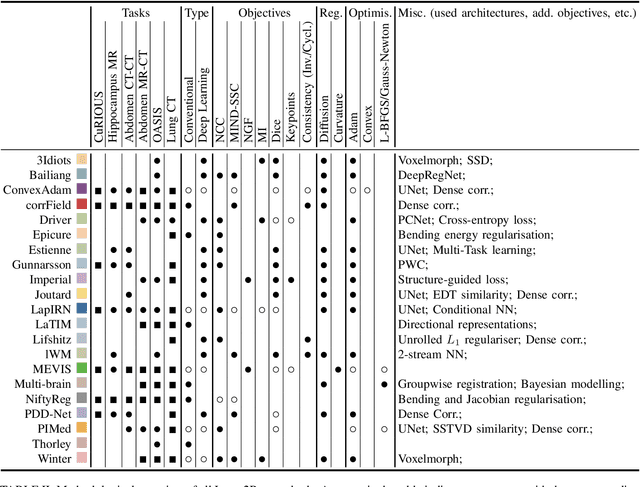
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge