Wayne G. Brisbane
Prostate Cancer Screening with Artificial Intelligence-Enhanced Micro-Ultrasound: A Comparative Study with Traditional Methods
May 27, 2025Abstract:Background and objective: Micro-ultrasound (micro-US) is a novel imaging modality with diagnostic accuracy comparable to MRI for detecting clinically significant prostate cancer (csPCa). We investigated whether artificial intelligence (AI) interpretation of micro-US can outperform clinical screening methods using PSA and digital rectal examination (DRE). Methods: We retrospectively studied 145 men who underwent micro-US guided biopsy (79 with csPCa, 66 without). A self-supervised convolutional autoencoder was used to extract deep image features from 2D micro-US slices. Random forest classifiers were trained using five-fold cross-validation to predict csPCa at the slice level. Patients were classified as csPCa-positive if 88 or more consecutive slices were predicted positive. Model performance was compared with a classifier using PSA, DRE, prostate volume, and age. Key findings and limitations: The AI-based micro-US model and clinical screening model achieved AUROCs of 0.871 and 0.753, respectively. At a fixed threshold, the micro-US model achieved 92.5% sensitivity and 68.1% specificity, while the clinical model showed 96.2% sensitivity but only 27.3% specificity. Limitations include a retrospective single-center design and lack of external validation. Conclusions and clinical implications: AI-interpreted micro-US improves specificity while maintaining high sensitivity for csPCa detection. This method may reduce unnecessary biopsies and serve as a low-cost alternative to PSA-based screening. Patient summary: We developed an AI system to analyze prostate micro-ultrasound images. It outperformed PSA and DRE in detecting aggressive cancer and may help avoid unnecessary biopsies.
Mask Enhanced Deeply Supervised Prostate Cancer Detection on B-mode Micro-Ultrasound
Dec 14, 2024



Abstract:Prostate cancer is a leading cause of cancer-related deaths among men. The recent development of high frequency, micro-ultrasound imaging offers improved resolution compared to conventional ultrasound and potentially a better ability to differentiate clinically significant cancer from normal tissue. However, the features of prostate cancer remain subtle, with ambiguous borders with normal tissue and large variations in appearance, making it challenging for both machine learning and humans to localize it on micro-ultrasound images. We propose a novel Mask Enhanced Deeply-supervised Micro-US network, termed MedMusNet, to automatically and more accurately segment prostate cancer to be used as potential targets for biopsy procedures. MedMusNet leverages predicted masks of prostate cancer to enforce the learned features layer-wisely within the network, reducing the influence of noise and improving overall consistency across frames. MedMusNet successfully detected 76% of clinically significant cancer with a Dice Similarity Coefficient of 0.365, significantly outperforming the baseline Swin-M2F in specificity and accuracy (Wilcoxon test, Bonferroni correction, p-value<0.05). While the lesion-level and patient-level analyses showed improved performance compared to human experts and different baseline, the improvements did not reach statistical significance, likely on account of the small cohort. We have presented a novel approach to automatically detect and segment clinically significant prostate cancer on B-mode micro-ultrasound images. Our MedMusNet model outperformed other models, surpassing even human experts. These preliminary results suggest the potential for aiding urologists in prostate cancer diagnosis via biopsy and treatment decision-making.
Image Registration of In Vivo Micro-Ultrasound and Ex Vivo Pseudo-Whole Mount Histopathology Images of the Prostate: A Proof-of-Concept Study
May 31, 2023



Abstract:Early diagnosis of prostate cancer significantly improves a patient's 5-year survival rate. Biopsy of small prostate cancers is improved with image-guided biopsy. MRI-ultrasound fusion-guided biopsy is sensitive to smaller tumors but is underutilized due to the high cost of MRI and fusion equipment. Micro-ultrasound (micro-US), a novel high-resolution ultrasound technology, provides a cost-effective alternative to MRI while delivering comparable diagnostic accuracy. However, the interpretation of micro-US is challenging due to subtle gray scale changes indicating cancer vs normal tissue. This challenge can be addressed by training urologists with a large dataset of micro-US images containing the ground truth cancer outlines. Such a dataset can be mapped from surgical specimens (histopathology) onto micro-US images via image registration. In this paper, we present a semi-automated pipeline for registering in vivo micro-US images with ex vivo whole-mount histopathology images. Our pipeline begins with the reconstruction of pseudo-whole-mount histopathology images and a 3D micro-US volume. Each pseudo-whole-mount histopathology image is then registered with the corresponding axial micro-US slice using a two-stage approach that estimates an affine transformation followed by a deformable transformation. We evaluated our registration pipeline using micro-US and histopathology images from 18 patients who underwent radical prostatectomy. The results showed a Dice coefficient of 0.94 and a landmark error of 2.7 mm, indicating the accuracy of our registration pipeline. This proof-of-concept study demonstrates the feasibility of accurately aligning micro-US and histopathology images. To promote transparency and collaboration in research, we will make our code and dataset publicly available.
MicroSegNet: A Deep Learning Approach for Prostate Segmentation on Micro-Ultrasound Images
May 31, 2023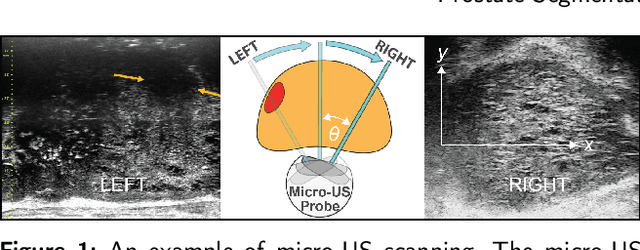
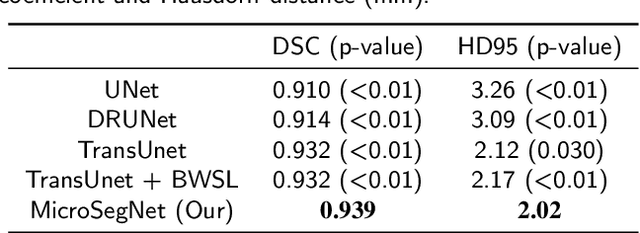
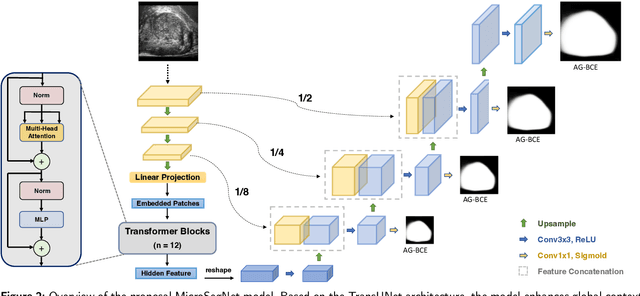
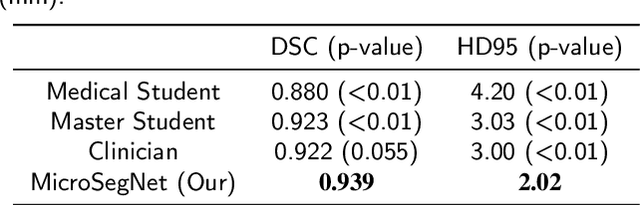
Abstract:Micro-ultrasound (micro-US) is a novel 29-MHz ultrasound technique that provides 3-4 times higher resolution than traditional ultrasound, delivering comparable accuracy for diagnosing prostate cancer to MRI but at a lower cost. Accurate prostate segmentation is crucial for prostate volume measurement, cancer diagnosis, prostate biopsy, and treatment planning. This paper proposes a deep learning approach for automated, fast, and accurate prostate segmentation on micro-US images. Prostate segmentation on micro-US is challenging due to artifacts and indistinct borders between the prostate, bladder, and urethra in the midline. We introduce MicroSegNet, a multi-scale annotation-guided Transformer UNet model to address this challenge. During the training process, MicroSegNet focuses more on regions that are hard to segment (challenging regions), where expert and non-expert annotations show discrepancies. We achieve this by proposing an annotation-guided cross entropy loss that assigns larger weight to pixels in hard regions and lower weight to pixels in easy regions. We trained our model using micro-US images from 55 patients, followed by evaluation on 20 patients. Our MicroSegNet model achieved a Dice coefficient of 0.942 and a Hausdorff distance of 2.11 mm, outperforming several state-of-the-art segmentation methods, as well as three human annotators with different experience levels. We will make our code and dataset publicly available to promote transparency and collaboration in research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge