Jake Pensa
Image Registration of In Vivo Micro-Ultrasound and Ex Vivo Pseudo-Whole Mount Histopathology Images of the Prostate: A Proof-of-Concept Study
May 31, 2023



Abstract:Early diagnosis of prostate cancer significantly improves a patient's 5-year survival rate. Biopsy of small prostate cancers is improved with image-guided biopsy. MRI-ultrasound fusion-guided biopsy is sensitive to smaller tumors but is underutilized due to the high cost of MRI and fusion equipment. Micro-ultrasound (micro-US), a novel high-resolution ultrasound technology, provides a cost-effective alternative to MRI while delivering comparable diagnostic accuracy. However, the interpretation of micro-US is challenging due to subtle gray scale changes indicating cancer vs normal tissue. This challenge can be addressed by training urologists with a large dataset of micro-US images containing the ground truth cancer outlines. Such a dataset can be mapped from surgical specimens (histopathology) onto micro-US images via image registration. In this paper, we present a semi-automated pipeline for registering in vivo micro-US images with ex vivo whole-mount histopathology images. Our pipeline begins with the reconstruction of pseudo-whole-mount histopathology images and a 3D micro-US volume. Each pseudo-whole-mount histopathology image is then registered with the corresponding axial micro-US slice using a two-stage approach that estimates an affine transformation followed by a deformable transformation. We evaluated our registration pipeline using micro-US and histopathology images from 18 patients who underwent radical prostatectomy. The results showed a Dice coefficient of 0.94 and a landmark error of 2.7 mm, indicating the accuracy of our registration pipeline. This proof-of-concept study demonstrates the feasibility of accurately aligning micro-US and histopathology images. To promote transparency and collaboration in research, we will make our code and dataset publicly available.
MicroSegNet: A Deep Learning Approach for Prostate Segmentation on Micro-Ultrasound Images
May 31, 2023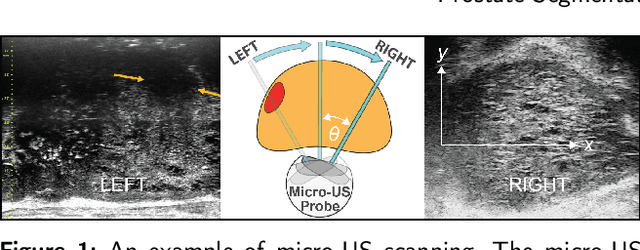
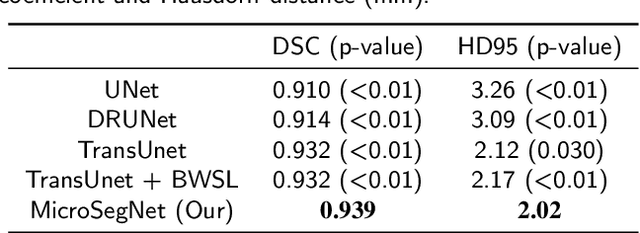
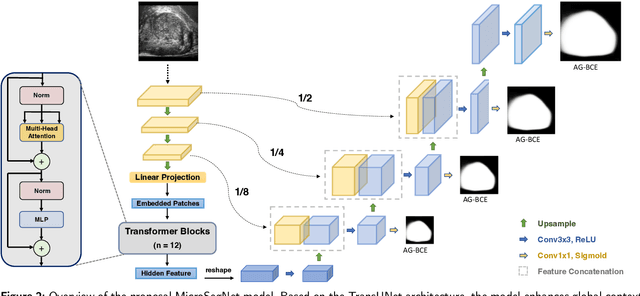
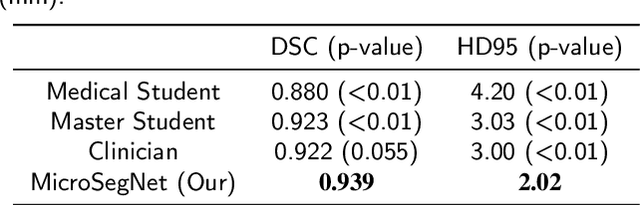
Abstract:Micro-ultrasound (micro-US) is a novel 29-MHz ultrasound technique that provides 3-4 times higher resolution than traditional ultrasound, delivering comparable accuracy for diagnosing prostate cancer to MRI but at a lower cost. Accurate prostate segmentation is crucial for prostate volume measurement, cancer diagnosis, prostate biopsy, and treatment planning. This paper proposes a deep learning approach for automated, fast, and accurate prostate segmentation on micro-US images. Prostate segmentation on micro-US is challenging due to artifacts and indistinct borders between the prostate, bladder, and urethra in the midline. We introduce MicroSegNet, a multi-scale annotation-guided Transformer UNet model to address this challenge. During the training process, MicroSegNet focuses more on regions that are hard to segment (challenging regions), where expert and non-expert annotations show discrepancies. We achieve this by proposing an annotation-guided cross entropy loss that assigns larger weight to pixels in hard regions and lower weight to pixels in easy regions. We trained our model using micro-US images from 55 patients, followed by evaluation on 20 patients. Our MicroSegNet model achieved a Dice coefficient of 0.942 and a Hausdorff distance of 2.11 mm, outperforming several state-of-the-art segmentation methods, as well as three human annotators with different experience levels. We will make our code and dataset publicly available to promote transparency and collaboration in research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge