Hongxu Jiang
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Fast-DDPM: Fast Denoising Diffusion Probabilistic Models for Medical Image-to-Image Generation
May 24, 2024



Abstract:Denoising diffusion probabilistic models (DDPMs) have achieved unprecedented success in computer vision. However, they remain underutilized in medical imaging, a field crucial for disease diagnosis and treatment planning. This is primarily due to the high computational cost associated with (1) the use of large number of time steps (e.g., 1,000) in diffusion processes and (2) the increased dimensionality of medical images, which are often 3D or 4D. Training a diffusion model on medical images typically takes days to weeks, while sampling each image volume takes minutes to hours. To address this challenge, we introduce Fast-DDPM, a simple yet effective approach capable of improving training speed, sampling speed, and generation quality simultaneously. Unlike DDPM, which trains the image denoiser across 1,000 time steps, Fast-DDPM trains and samples using only 10 time steps. The key to our method lies in aligning the training and sampling procedures to optimize time-step utilization. Specifically, we introduced two efficient noise schedulers with 10 time steps: one with uniform time step sampling and another with non-uniform sampling. We evaluated Fast-DDPM across three medical image-to-image generation tasks: multi-image super-resolution, image denoising, and image-to-image translation. Fast-DDPM outperformed DDPM and current state-of-the-art methods based on convolutional networks and generative adversarial networks in all tasks. Additionally, Fast-DDPM reduced the training time to 0.2x and the sampling time to 0.01x compared to DDPM. Our code is publicly available at: https://github.com/mirthAI/Fast-DDPM.
Fast Denoising Diffusion Probabilistic Models for Medical Image-to-Image Generation
May 23, 2024



Abstract:Denoising diffusion probabilistic models (DDPMs) have achieved unprecedented success in computer vision. However, they remain underutilized in medical imaging, a field crucial for disease diagnosis and treatment planning. This is primarily due to the high computational cost associated with (1) the use of large number of time steps (e.g., 1,000) in diffusion processes and (2) the increased dimensionality of medical images, which are often 3D or 4D. Training a diffusion model on medical images typically takes days to weeks, while sampling each image volume takes minutes to hours. To address this challenge, we introduce Fast-DDPM, a simple yet effective approach capable of improving training speed, sampling speed, and generation quality simultaneously. Unlike DDPM, which trains the image denoiser across 1,000 time steps, Fast-DDPM trains and samples using only 10 time steps. The key to our method lies in aligning the training and sampling procedures. We introduced two efficient noise schedulers with 10 time steps: one with uniform time step sampling and another with non-uniform sampling. We evaluated Fast-DDPM across three medical image-to-image generation tasks: multi-image super-resolution, image denoising, and image-to-image translation. Fast-DDPM outperformed DDPM and current state-of-the-art methods based on convolutional networks and generative adversarial networks in all tasks. Additionally, Fast-DDPM reduced training time by a factor of 5 and sampling time by a factor of 100 compared to DDPM. Our code is publicly available at: https://github.com/mirthAI/Fast-DDPM.
A Flexible 2.5D Medical Image Segmentation Approach with In-Slice and Cross-Slice Attention
Apr 30, 2024
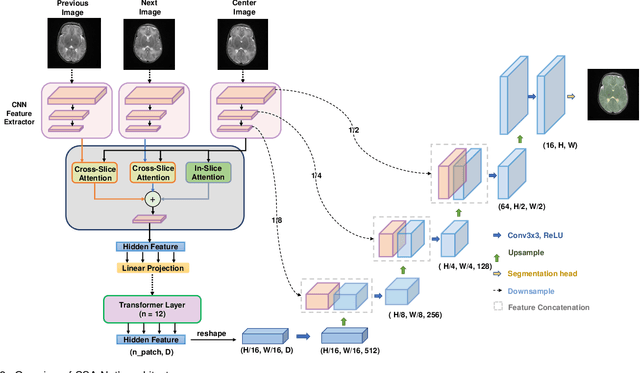
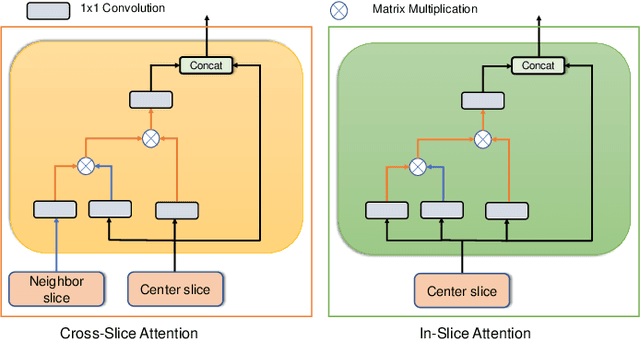
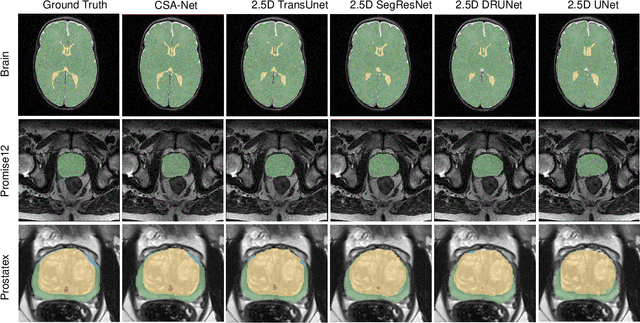
Abstract:Deep learning has become the de facto method for medical image segmentation, with 3D segmentation models excelling in capturing complex 3D structures and 2D models offering high computational efficiency. However, segmenting 2.5D images, which have high in-plane but low through-plane resolution, is a relatively unexplored challenge. While applying 2D models to individual slices of a 2.5D image is feasible, it fails to capture the spatial relationships between slices. On the other hand, 3D models face challenges such as resolution inconsistencies in 2.5D images, along with computational complexity and susceptibility to overfitting when trained with limited data. In this context, 2.5D models, which capture inter-slice correlations using only 2D neural networks, emerge as a promising solution due to their reduced computational demand and simplicity in implementation. In this paper, we introduce CSA-Net, a flexible 2.5D segmentation model capable of processing 2.5D images with an arbitrary number of slices through an innovative Cross-Slice Attention (CSA) module. This module uses the cross-slice attention mechanism to effectively capture 3D spatial information by learning long-range dependencies between the center slice (for segmentation) and its neighboring slices. Moreover, CSA-Net utilizes the self-attention mechanism to understand correlations among pixels within the center slice. We evaluated CSA-Net on three 2.5D segmentation tasks: (1) multi-class brain MRI segmentation, (2) binary prostate MRI segmentation, and (3) multi-class prostate MRI segmentation. CSA-Net outperformed leading 2D and 2.5D segmentation methods across all three tasks, demonstrating its efficacy and superiority. Our code is publicly available at https://github.com/mirthAI/CSA-Net.
SAM-Lightening: A Lightweight Segment Anything Model with Dilated Flash Attention to Achieve 30 times Acceleration
Mar 18, 2024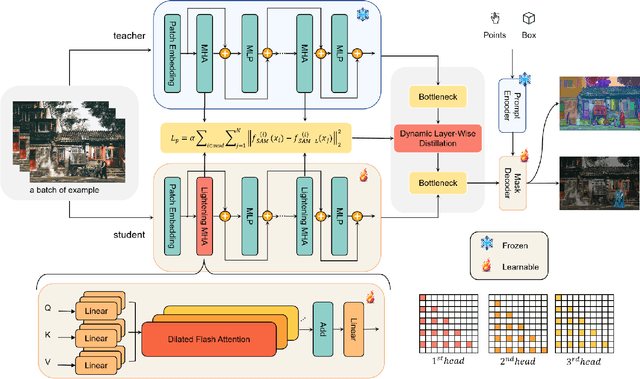
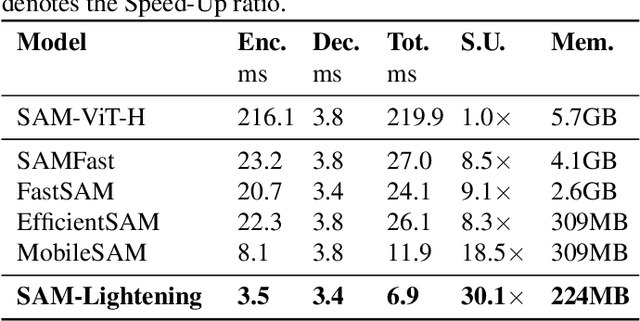

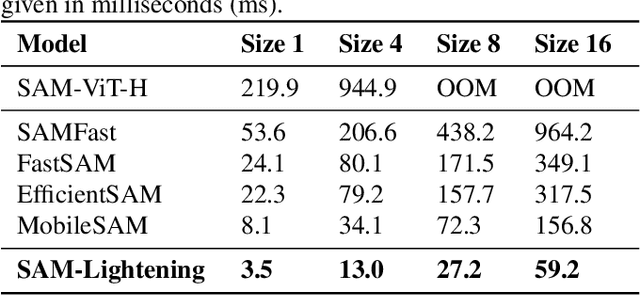
Abstract:Segment Anything Model (SAM) has garnered significant attention in segmentation tasks due to their zero-shot generalization ability. However, a broader application of SAMs to real-world practice has been restricted by their low inference speed and high computational memory demands, which mainly stem from the attention mechanism. Existing work concentrated on optimizing the encoder, yet has not adequately addressed the inefficiency of the attention mechanism itself, even when distilled to a smaller model, which thus leaves space for further improvement. In response, we introduce SAM-Lightening, a variant of SAM, that features a re-engineered attention mechanism, termed Dilated Flash Attention. It not only facilitates higher parallelism, enhancing processing efficiency but also retains compatibility with the existing FlashAttention. Correspondingly, we propose a progressive distillation to enable an efficient knowledge transfer from the vanilla SAM without costly training from scratch. Experiments on COCO and LVIS reveal that SAM-Lightening significantly outperforms the state-of-the-art methods in both run-time efficiency and segmentation accuracy. Specifically, it can achieve an inference speed of 7 milliseconds (ms) per image, for images of size 1024*1024 pixels, which is 30.1 times faster than the vanilla SAM and 2.1 times than the state-of-the-art. Moreover, it takes only 244MB memory, which is 3.5\% of the vanilla SAM. The code and weights are available at https://anonymous.4open.science/r/SAM-LIGHTENING-BC25/.
MicroSegNet: A Deep Learning Approach for Prostate Segmentation on Micro-Ultrasound Images
May 31, 2023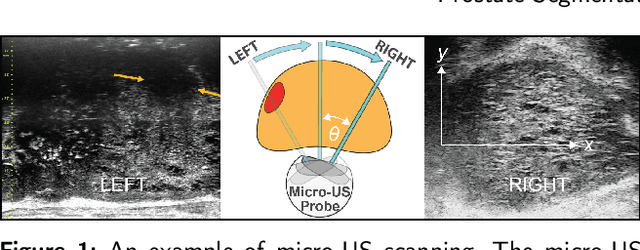
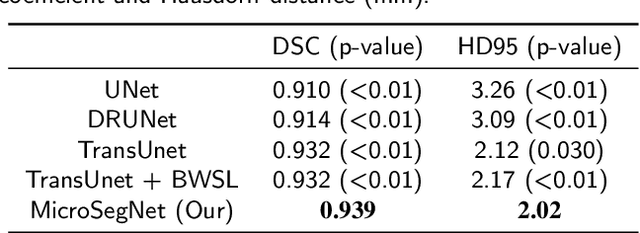
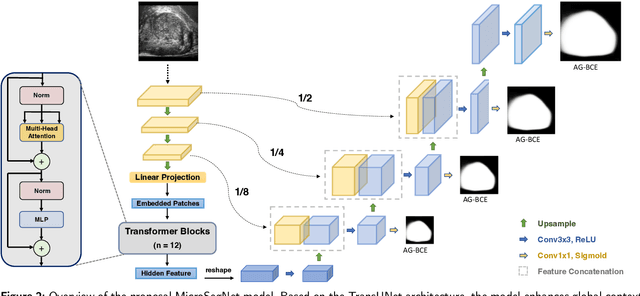
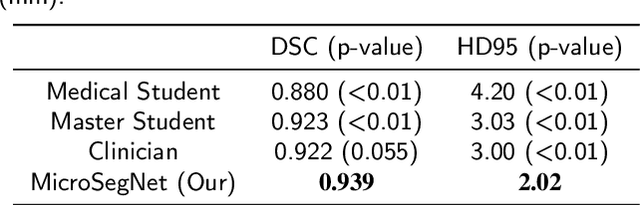
Abstract:Micro-ultrasound (micro-US) is a novel 29-MHz ultrasound technique that provides 3-4 times higher resolution than traditional ultrasound, delivering comparable accuracy for diagnosing prostate cancer to MRI but at a lower cost. Accurate prostate segmentation is crucial for prostate volume measurement, cancer diagnosis, prostate biopsy, and treatment planning. This paper proposes a deep learning approach for automated, fast, and accurate prostate segmentation on micro-US images. Prostate segmentation on micro-US is challenging due to artifacts and indistinct borders between the prostate, bladder, and urethra in the midline. We introduce MicroSegNet, a multi-scale annotation-guided Transformer UNet model to address this challenge. During the training process, MicroSegNet focuses more on regions that are hard to segment (challenging regions), where expert and non-expert annotations show discrepancies. We achieve this by proposing an annotation-guided cross entropy loss that assigns larger weight to pixels in hard regions and lower weight to pixels in easy regions. We trained our model using micro-US images from 55 patients, followed by evaluation on 20 patients. Our MicroSegNet model achieved a Dice coefficient of 0.942 and a Hausdorff distance of 2.11 mm, outperforming several state-of-the-art segmentation methods, as well as three human annotators with different experience levels. We will make our code and dataset publicly available to promote transparency and collaboration in research.
Human-centric Spatio-Temporal Video Grounding With Visual Transformers
Nov 10, 2020

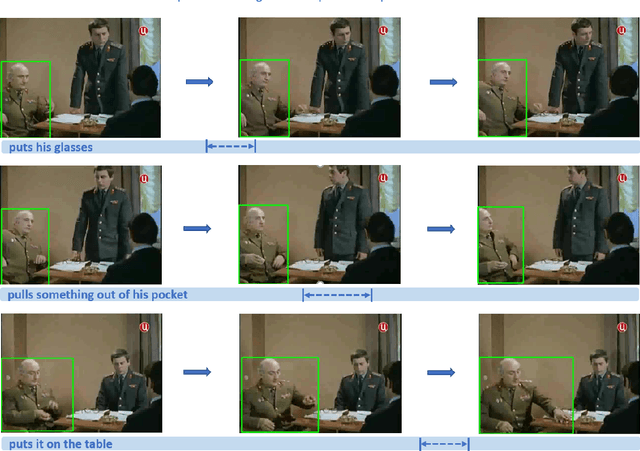
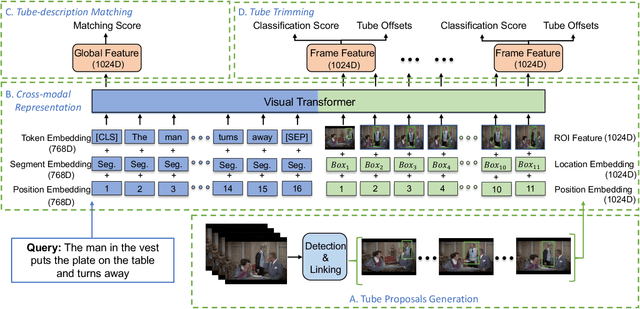
Abstract:In this work, we introduce a novel task - Humancentric Spatio-Temporal Video Grounding (HC-STVG). Unlike the existing referring expression tasks in images or videos, by focusing on humans, HC-STVG aims to localize a spatiotemporal tube of the target person from an untrimmed video based on a given textural description. This task is useful, especially for healthcare and security-related applications, where the surveillance videos can be extremely long but only a specific person during a specific period of time is concerned. HC-STVG is a video grounding task that requires both spatial (where) and temporal (when) localization. Unfortunately, the existing grounding methods cannot handle this task well. We tackle this task by proposing an effective baseline method named Spatio-Temporal Grounding with Visual Transformers (STGVT), which utilizes Visual Transformers to extract cross-modal representations for video-sentence matching and temporal localization. To facilitate this task, we also contribute an HC-STVG dataset consisting of 5,660 video-sentence pairs on complex multi-person scenes. Specifically, each video lasts for 20 seconds, pairing with a natural query sentence with an average of 17.25 words. Extensive experiments are conducted on this dataset, demonstrating the newly-proposed method outperforms the existing baseline methods.
Robustness-aware 2-bit quantization with real-time performance for neural network
Oct 19, 2020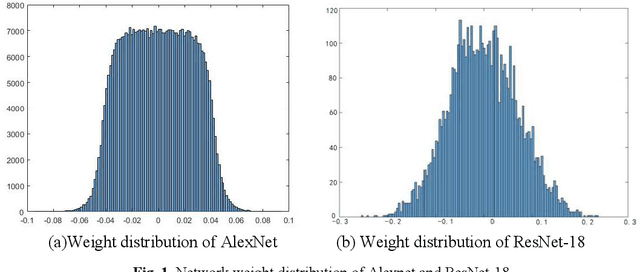
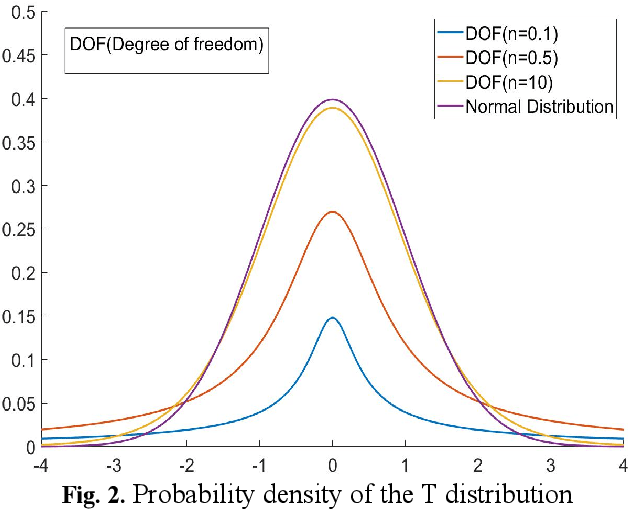
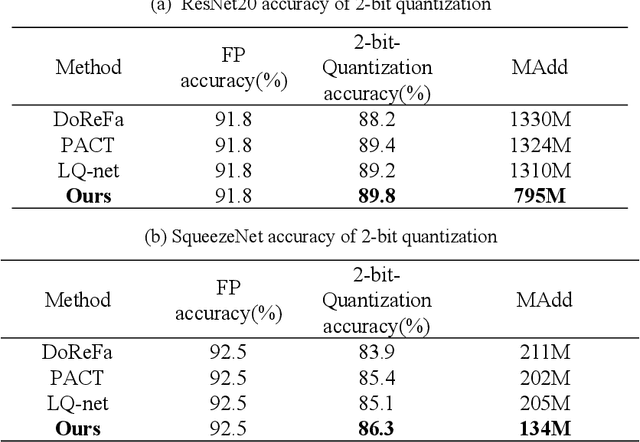
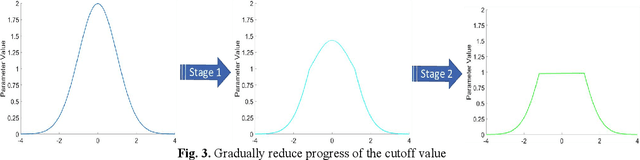
Abstract:Quantized neural network (NN) with a reduced bit precision is an effective solution to reduces the computational and memory resource requirements and plays a vital role in machine learning. However, it is still challenging to avoid the significant accuracy degradation due to its numerical approximation and lower redundancy. In this paper, a novel robustness-aware 2-bit quantization scheme is proposed for NN base on binary NN and generative adversarial network(GAN), witch improves the performance by enriching the information of binary NN, efficiently extract the structural information and considering the robustness of the quantized NN. Specifically, using shift addition operation to replace the multiply-accumulate in the quantization process witch can effectively speed the NN. Meanwhile, a structural loss between the original NN and quantized NN is proposed to such that the structural information of data is preserved after quantization. The structural information learned from NN not only plays an important role in improving the performance but also allows for further fine tuning of the quantization network by applying the Lipschitz constraint to the structural loss. In addition, we also for the first time take the robustness of the quantized NN into consideration and propose a non-sensitive perturbation loss function by introducing an extraneous term of spectral norm. The experiments are conducted on CIFAR-10 and ImageNet datasets with popular NN( such as MoblieNetV2, SqueezeNet, ResNet20, etc). The experimental results show that the proposed algorithm is more competitive under 2-bit-precision than the state-of-the-art quantization methods. Meanwhile, the experimental results also demonstrate that the proposed method is robust under the FGSM adversarial samples attack.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge