Muxuan Liang
Uncertainty-Aware Prediction of Parkinson's Disease Medication Needs: A Two-Stage Conformal Prediction Approach
Aug 14, 2025Abstract:Parkinson's Disease (PD) medication management presents unique challenges due to heterogeneous disease progression and treatment response. Neurologists must balance symptom control with optimal dopaminergic dosing based on functional disability while minimizing side effects. This balance is crucial as inadequate or abrupt changes can cause levodopa-induced dyskinesia, wearing off, and neuropsychiatric effects, significantly reducing quality of life. Current approaches rely on trial-and-error decisions without systematic predictive methods. Despite machine learning advances, clinical adoption remains limited due to reliance on point predictions that do not account for prediction uncertainty, undermining clinical trust and utility. Clinicians require not only predictions of future medication needs but also reliable confidence measures. Without quantified uncertainty, adjustments risk premature escalation to maximum doses or prolonged inadequate symptom control. We developed a conformal prediction framework anticipating medication needs up to two years in advance with reliable prediction intervals and statistical guarantees. Our approach addresses zero-inflation in PD inpatient data, where patients maintain stable medication regimens between visits. Using electronic health records from 631 inpatient admissions at University of Florida Health (2011-2021), our two-stage approach identifies patients likely to need medication changes, then predicts required levodopa equivalent daily dose adjustments. Our framework achieved marginal coverage while reducing prediction interval lengths compared to traditional approaches, providing precise predictions for short-term planning and wider ranges for long-term forecasting. By quantifying uncertainty, our approach enables evidence-based decisions about levodopa dosing, optimizing symptom control while minimizing side effects and improving life quality.
Late Fusion Multi-task Learning for Semiparametric Inference with Nuisance Parameters
Jul 10, 2025Abstract:In the age of large and heterogeneous datasets, the integration of information from diverse sources is essential to improve parameter estimation. Multi-task learning offers a powerful approach by enabling simultaneous learning across related tasks. In this work, we introduce a late fusion framework for multi-task learning with semiparametric models that involve infinite-dimensional nuisance parameters, focusing on applications such as heterogeneous treatment effect estimation across multiple data sources, including electronic health records from different hospitals or clinical trial data. Our framework is two-step: first, initial double machine-learning estimators are obtained through individual task learning; second, these estimators are adaptively aggregated to exploit task similarities while remaining robust to task-specific differences. In particular, the framework avoids individual level data sharing, preserving privacy. Additionally, we propose a novel multi-task learning method for nuisance parameter estimation, which further enhances parameter estimation when nuisance parameters exhibit similarity across tasks. We establish theoretical guarantees for the method, demonstrating faster convergence rates compared to individual task learning when tasks share similar parametric components. Extensive simulations and real data applications complement the theoretical findings of our work while highlight the effectiveness of our framework even in moderate sample sizes.
Multi-Class Segmentation of Aortic Branches and Zones in Computed Tomography Angiography: The AortaSeg24 Challenge
Feb 07, 2025



Abstract:Multi-class segmentation of the aorta in computed tomography angiography (CTA) scans is essential for diagnosing and planning complex endovascular treatments for patients with aortic dissections. However, existing methods reduce aortic segmentation to a binary problem, limiting their ability to measure diameters across different branches and zones. Furthermore, no open-source dataset is currently available to support the development of multi-class aortic segmentation methods. To address this gap, we organized the AortaSeg24 MICCAI Challenge, introducing the first dataset of 100 CTA volumes annotated for 23 clinically relevant aortic branches and zones. This dataset was designed to facilitate both model development and validation. The challenge attracted 121 teams worldwide, with participants leveraging state-of-the-art frameworks such as nnU-Net and exploring novel techniques, including cascaded models, data augmentation strategies, and custom loss functions. We evaluated the submitted algorithms using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD), highlighting the approaches adopted by the top five performing teams. This paper presents the challenge design, dataset details, evaluation metrics, and an in-depth analysis of the top-performing algorithms. The annotated dataset, evaluation code, and implementations of the leading methods are publicly available to support further research. All resources can be accessed at https://aortaseg24.grand-challenge.org.
Fast-DDPM: Fast Denoising Diffusion Probabilistic Models for Medical Image-to-Image Generation
May 24, 2024



Abstract:Denoising diffusion probabilistic models (DDPMs) have achieved unprecedented success in computer vision. However, they remain underutilized in medical imaging, a field crucial for disease diagnosis and treatment planning. This is primarily due to the high computational cost associated with (1) the use of large number of time steps (e.g., 1,000) in diffusion processes and (2) the increased dimensionality of medical images, which are often 3D or 4D. Training a diffusion model on medical images typically takes days to weeks, while sampling each image volume takes minutes to hours. To address this challenge, we introduce Fast-DDPM, a simple yet effective approach capable of improving training speed, sampling speed, and generation quality simultaneously. Unlike DDPM, which trains the image denoiser across 1,000 time steps, Fast-DDPM trains and samples using only 10 time steps. The key to our method lies in aligning the training and sampling procedures to optimize time-step utilization. Specifically, we introduced two efficient noise schedulers with 10 time steps: one with uniform time step sampling and another with non-uniform sampling. We evaluated Fast-DDPM across three medical image-to-image generation tasks: multi-image super-resolution, image denoising, and image-to-image translation. Fast-DDPM outperformed DDPM and current state-of-the-art methods based on convolutional networks and generative adversarial networks in all tasks. Additionally, Fast-DDPM reduced the training time to 0.2x and the sampling time to 0.01x compared to DDPM. Our code is publicly available at: https://github.com/mirthAI/Fast-DDPM.
Fast Denoising Diffusion Probabilistic Models for Medical Image-to-Image Generation
May 23, 2024



Abstract:Denoising diffusion probabilistic models (DDPMs) have achieved unprecedented success in computer vision. However, they remain underutilized in medical imaging, a field crucial for disease diagnosis and treatment planning. This is primarily due to the high computational cost associated with (1) the use of large number of time steps (e.g., 1,000) in diffusion processes and (2) the increased dimensionality of medical images, which are often 3D or 4D. Training a diffusion model on medical images typically takes days to weeks, while sampling each image volume takes minutes to hours. To address this challenge, we introduce Fast-DDPM, a simple yet effective approach capable of improving training speed, sampling speed, and generation quality simultaneously. Unlike DDPM, which trains the image denoiser across 1,000 time steps, Fast-DDPM trains and samples using only 10 time steps. The key to our method lies in aligning the training and sampling procedures. We introduced two efficient noise schedulers with 10 time steps: one with uniform time step sampling and another with non-uniform sampling. We evaluated Fast-DDPM across three medical image-to-image generation tasks: multi-image super-resolution, image denoising, and image-to-image translation. Fast-DDPM outperformed DDPM and current state-of-the-art methods based on convolutional networks and generative adversarial networks in all tasks. Additionally, Fast-DDPM reduced training time by a factor of 5 and sampling time by a factor of 100 compared to DDPM. Our code is publicly available at: https://github.com/mirthAI/Fast-DDPM.
Inference with non-differentiable surrogate loss in a general high-dimensional classification framework
May 20, 2024Abstract:Penalized empirical risk minimization with a surrogate loss function is often used to derive a high-dimensional linear decision rule in classification problems. Although much of the literature focuses on the generalization error, there is a lack of valid inference procedures to identify the driving factors of the estimated decision rule, especially when the surrogate loss is non-differentiable. In this work, we propose a kernel-smoothed decorrelated score to construct hypothesis testing and interval estimations for the linear decision rule estimated using a piece-wise linear surrogate loss, which has a discontinuous gradient and non-regular Hessian. Specifically, we adopt kernel approximations to smooth the discontinuous gradient near discontinuity points and approximate the non-regular Hessian of the surrogate loss. In applications where additional nuisance parameters are involved, we propose a novel cross-fitted version to accommodate flexible nuisance estimates and kernel approximations. We establish the limiting distribution of the kernel-smoothed decorrelated score and its cross-fitted version in a high-dimensional setup. Simulation and real data analysis are conducted to demonstrate the validity and superiority of the proposed method.
MicroSegNet: A Deep Learning Approach for Prostate Segmentation on Micro-Ultrasound Images
May 31, 2023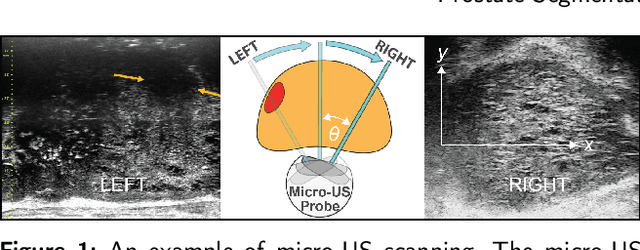
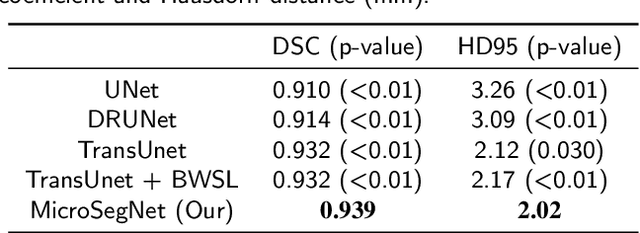
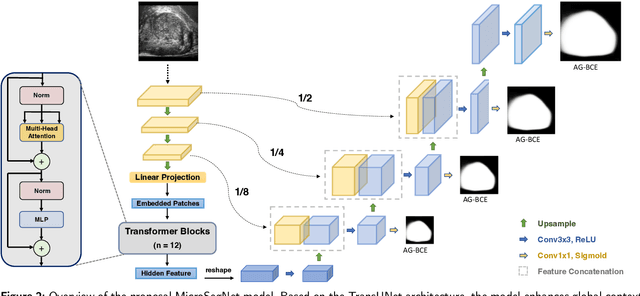
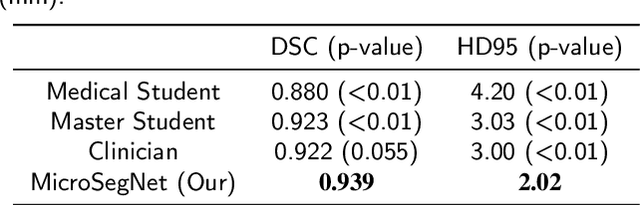
Abstract:Micro-ultrasound (micro-US) is a novel 29-MHz ultrasound technique that provides 3-4 times higher resolution than traditional ultrasound, delivering comparable accuracy for diagnosing prostate cancer to MRI but at a lower cost. Accurate prostate segmentation is crucial for prostate volume measurement, cancer diagnosis, prostate biopsy, and treatment planning. This paper proposes a deep learning approach for automated, fast, and accurate prostate segmentation on micro-US images. Prostate segmentation on micro-US is challenging due to artifacts and indistinct borders between the prostate, bladder, and urethra in the midline. We introduce MicroSegNet, a multi-scale annotation-guided Transformer UNet model to address this challenge. During the training process, MicroSegNet focuses more on regions that are hard to segment (challenging regions), where expert and non-expert annotations show discrepancies. We achieve this by proposing an annotation-guided cross entropy loss that assigns larger weight to pixels in hard regions and lower weight to pixels in easy regions. We trained our model using micro-US images from 55 patients, followed by evaluation on 20 patients. Our MicroSegNet model achieved a Dice coefficient of 0.942 and a Hausdorff distance of 2.11 mm, outperforming several state-of-the-art segmentation methods, as well as three human annotators with different experience levels. We will make our code and dataset publicly available to promote transparency and collaboration in research.
Learning a high-dimensional classification rule using auxiliary outcomes
Nov 11, 2020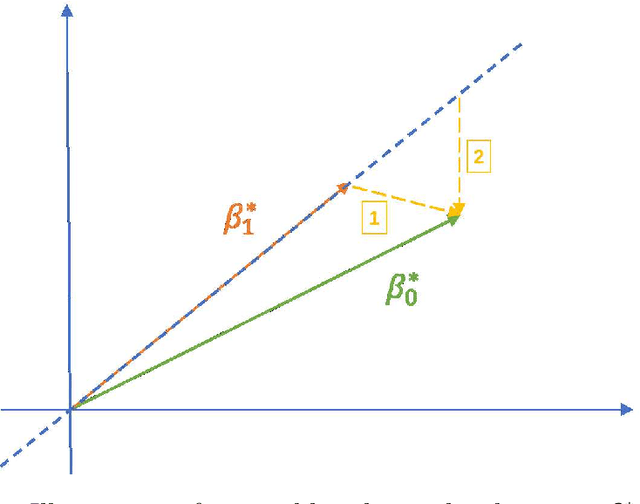
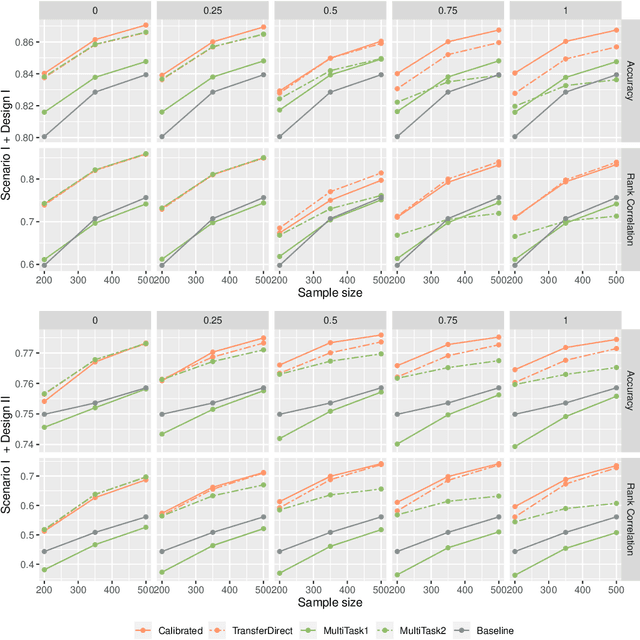
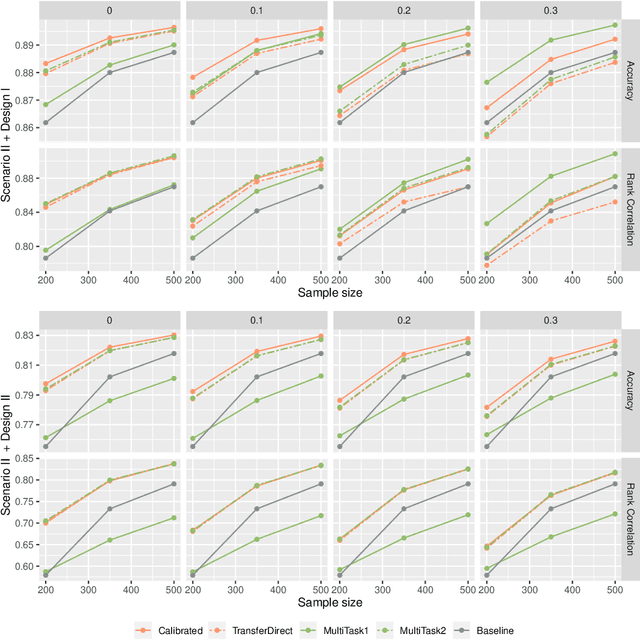
Abstract:Correlated outcomes are common in many practical problems. Based on a decomposition of estimation bias into two types, within-subspace and against-subspace, we develop a robust approach to estimating the classification rule for the outcome of interest with the presence of auxiliary outcomes in high-dimensional settings. The proposed method includes a pooled estimation step using all outcomes to gain efficiency, and a subsequent calibration step using only the outcome of interest to correct both types of biases. We show that when the pooled estimator has a low estimation error and a sparse against-subspace bias, the calibrated estimator can achieve a lower estimation error than that when using only the single outcome of interest. An inference procedure for the calibrated estimator is also provided. Simulations and a real data analysis are conducted to justify the superiority of the proposed method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge