Kuang Gong
for the Alzheimer's Disease Neuroimaging Initiative
MedVL-SAM2: A unified 3D medical vision-language model for multimodal reasoning and prompt-driven segmentation
Jan 14, 2026Abstract:Recent progress in medical vision-language models (VLMs) has achieved strong performance on image-level text-centric tasks such as report generation and visual question answering (VQA). However, achieving fine-grained visual grounding and volumetric spatial reasoning in 3D medical VLMs remains challenging, particularly when aiming to unify these capabilities within a single, generalizable framework. To address this challenge, we proposed MedVL-SAM2, a unified 3D medical multimodal model that concurrently supports report generation, VQA, and multi-paradigm segmentation, including semantic, referring, and interactive segmentation. MedVL-SAM2 integrates image-level reasoning and pixel-level perception through a cohesive architecture tailored for 3D medical imaging, and incorporates a SAM2-based volumetric segmentation module to enable precise multi-granular spatial reasoning. The model is trained in a multi-stage pipeline: it is first pre-trained on a large-scale corpus of 3D CT image-text pairs to align volumetric visual features with radiology-language embeddings. It is then jointly optimized with both language-understanding and segmentation objectives using a comprehensive 3D CT segmentation dataset. This joint training enables flexible interaction via language, point, or box prompts, thereby unifying high-level visual reasoning with spatially precise localization. Our unified architecture delivers state-of-the-art performance across report generation, VQA, and multiple 3D segmentation tasks. Extensive analyses further show that the model provides reliable 3D visual grounding, controllable interactive segmentation, and robust cross-modal reasoning, demonstrating that high-level semantic reasoning and precise 3D localization can be jointly achieved within a unified 3D medical VLM.
Patlak Parametric Image Estimation from Dynamic PET Using Diffusion Model Prior
Dec 22, 2025Abstract:Dynamic PET enables the quantitative estimation of physiology-related parameters and is widely utilized in research and increasingly adopted in clinical settings. Parametric imaging in dynamic PET requires kinetic modeling to estimate voxel-wise physiological parameters based on specific kinetic models. However, parametric images estimated through kinetic model fitting often suffer from low image quality due to the inherently ill-posed nature of the fitting process and the limited counts resulting from non-continuous data acquisition across multiple bed positions in whole-body PET. In this work, we proposed a diffusion model-based kinetic modeling framework for parametric image estimation, using the Patlak model as an example. The score function of the diffusion model was pre-trained on static total-body PET images and served as a prior for both Patlak slope and intercept images by leveraging their patch-wise similarity. During inference, the kinetic model was incorporated as a data-consistency constraint to guide the parametric image estimation. The proposed framework was evaluated on total-body dynamic PET datasets with different dose levels, demonstrating the feasibility and promising performance of the proposed framework in improving parametric image quality.
TauGenNet: Plasma-Driven Tau PET Image Synthesis via Text-Guided 3D Diffusion Models
Sep 04, 2025



Abstract:Accurate quantification of tau pathology via tau positron emission tomography (PET) scan is crucial for diagnosing and monitoring Alzheimer's disease (AD). However, the high cost and limited availability of tau PET restrict its widespread use. In contrast, structural magnetic resonance imaging (MRI) and plasma-based biomarkers provide non-invasive and widely available complementary information related to brain anatomy and disease progression. In this work, we propose a text-guided 3D diffusion model for 3D tau PET image synthesis, leveraging multimodal conditions from both structural MRI and plasma measurement. Specifically, the textual prompt is from the plasma p-tau217 measurement, which is a key indicator of AD progression, while MRI provides anatomical structure constraints. The proposed framework is trained and evaluated using clinical AV1451 tau PET data from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database. Experimental results demonstrate that our approach can generate realistic, clinically meaningful 3D tau PET across a range of disease stages. The proposed framework can help perform tau PET data augmentation under different settings, provide a non-invasive, cost-effective alternative for visualizing tau pathology, and support the simulation of disease progression under varying plasma biomarker levels and cognitive conditions.
SAM2-SGP: Enhancing SAM2 for Medical Image Segmentation via Support-Set Guided Prompting
Jun 24, 2025



Abstract:Although new vision foundation models such as Segment Anything Model 2 (SAM2) have significantly enhanced zero-shot image segmentation capabilities, reliance on human-provided prompts poses significant challenges in adapting SAM2 to medical image segmentation tasks. Moreover, SAM2's performance in medical image segmentation was limited by the domain shift issue, since it was originally trained on natural images and videos. To address these challenges, we proposed SAM2 with support-set guided prompting (SAM2-SGP), a framework that eliminated the need for manual prompts. The proposed model leveraged the memory mechanism of SAM2 to generate pseudo-masks using image-mask pairs from a support set via a Pseudo-mask Generation (PMG) module. We further introduced a novel Pseudo-mask Attention (PMA) module, which used these pseudo-masks to automatically generate bounding boxes and enhance localized feature extraction by guiding attention to relevant areas. Furthermore, a low-rank adaptation (LoRA) strategy was adopted to mitigate the domain shift issue. The proposed framework was evaluated on both 2D and 3D datasets across multiple medical imaging modalities, including fundus photography, X-ray, computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), and ultrasound. The results demonstrated a significant performance improvement over state-of-the-art models, such as nnUNet and SwinUNet, as well as foundation models, such as SAM2 and MedSAM2, underscoring the effectiveness of the proposed approach. Our code is publicly available at https://github.com/astlian9/SAM_Support.
CDPDNet: Integrating Text Guidance with Hybrid Vision Encoders for Medical Image Segmentation
May 25, 2025



Abstract:Most publicly available medical segmentation datasets are only partially labeled, with annotations provided for a subset of anatomical structures. When multiple datasets are combined for training, this incomplete annotation poses challenges, as it limits the model's ability to learn shared anatomical representations among datasets. Furthermore, vision-only frameworks often fail to capture complex anatomical relationships and task-specific distinctions, leading to reduced segmentation accuracy and poor generalizability to unseen datasets. In this study, we proposed a novel CLIP-DINO Prompt-Driven Segmentation Network (CDPDNet), which combined a self-supervised vision transformer with CLIP-based text embedding and introduced task-specific text prompts to tackle these challenges. Specifically, the framework was constructed upon a convolutional neural network (CNN) and incorporated DINOv2 to extract both fine-grained and global visual features, which were then fused using a multi-head cross-attention module to overcome the limited long-range modeling capability of CNNs. In addition, CLIP-derived text embeddings were projected into the visual space to help model complex relationships among organs and tumors. To further address the partial label challenge and enhance inter-task discriminative capability, a Text-based Task Prompt Generation (TTPG) module that generated task-specific prompts was designed to guide the segmentation. Extensive experiments on multiple medical imaging datasets demonstrated that CDPDNet consistently outperformed existing state-of-the-art segmentation methods. Code and pretrained model are available at: https://github.com/wujiong-hub/CDPDNet.git.
Med3DVLM: An Efficient Vision-Language Model for 3D Medical Image Analysis
Mar 25, 2025Abstract:Vision-language models (VLMs) have shown promise in 2D medical image analysis, but extending them to 3D remains challenging due to the high computational demands of volumetric data and the difficulty of aligning 3D spatial features with clinical text. We present Med3DVLM, a 3D VLM designed to address these challenges through three key innovations: (1) DCFormer, an efficient encoder that uses decomposed 3D convolutions to capture fine-grained spatial features at scale; (2) SigLIP, a contrastive learning strategy with pairwise sigmoid loss that improves image-text alignment without relying on large negative batches; and (3) a dual-stream MLP-Mixer projector that fuses low- and high-level image features with text embeddings for richer multi-modal representations. We evaluate our model on the M3D dataset, which includes radiology reports and VQA data for 120,084 3D medical images. Results show that Med3DVLM achieves superior performance across multiple benchmarks. For image-text retrieval, it reaches 61.00% R@1 on 2,000 samples, significantly outperforming the current state-of-the-art M3D model (19.10%). For report generation, it achieves a METEOR score of 36.42% (vs. 14.38%). In open-ended visual question answering (VQA), it scores 36.76% METEOR (vs. 33.58%), and in closed-ended VQA, it achieves 79.95% accuracy (vs. 75.78%). These results highlight Med3DVLM's ability to bridge the gap between 3D imaging and language, enabling scalable, multi-task reasoning across clinical applications. Our code is publicly available at https://github.com/mirthAI/Med3DVLM.
PET Image Denoising via Text-Guided Diffusion: Integrating Anatomical Priors through Text Prompts
Feb 28, 2025Abstract:Low-dose Positron Emission Tomography (PET) imaging presents a significant challenge due to increased noise and reduced image quality, which can compromise its diagnostic accuracy and clinical utility. Denoising diffusion probabilistic models (DDPMs) have demonstrated promising performance for PET image denoising. However, existing DDPM-based methods typically overlook valuable metadata such as patient demographics, anatomical information, and scanning parameters, which should further enhance the denoising performance if considered. Recent advances in vision-language models (VLMs), particularly the pre-trained Contrastive Language-Image Pre-training (CLIP) model, have highlighted the potential of incorporating text-based information into visual tasks to improve downstream performance. In this preliminary study, we proposed a novel text-guided DDPM for PET image denoising that integrated anatomical priors through text prompts. Anatomical text descriptions were encoded using a pre-trained CLIP text encoder to extract semantic guidance, which was then incorporated into the diffusion process via the cross-attention mechanism. Evaluations based on paired 1/20 low-dose and normal-dose 18F-FDG PET datasets demonstrated that the proposed method achieved better quantitative performance than conventional UNet and standard DDPM methods at both the whole-body and organ levels. These results underscored the potential of leveraging VLMs to integrate rich metadata into the diffusion framework to enhance the image quality of low-dose PET scans.
DCFormer: Efficient 3D Vision-Language Modeling with Decomposed Convolutions
Feb 07, 2025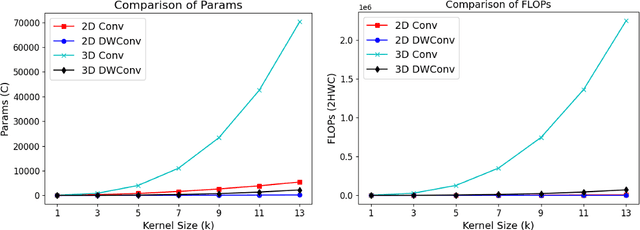



Abstract:Vision-language models (VLMs) align visual and textual representations, enabling high-performance zero-shot classification and image-text retrieval in 2D medical imaging. However, extending VLMs to 3D medical imaging remains computationally challenging. Existing 3D VLMs rely on Vision Transformers (ViTs), which are computationally expensive due to self-attention's quadratic complexity, or 3D convolutions, which demand excessive parameters and FLOPs as kernel size increases. We introduce DCFormer, an efficient 3D medical image encoder that factorizes 3D convolutions into three parallel 1D convolutions along depth, height, and width. This design preserves spatial information while significantly reducing computational cost. Integrated into a CLIP-based vision-language framework, DCFormer is evaluated on CT-RATE, a dataset of 50,188 paired 3D chest CT volumes and radiology reports, for zero-shot multi-abnormality detection across 18 pathologies. Compared to ViT, ConvNeXt, PoolFormer, and TransUNet, DCFormer achieves superior efficiency and accuracy, with DCFormer-Tiny reaching 62.0% accuracy and a 46.3% F1-score while using significantly fewer parameters. These results highlight DCFormer's potential for scalable, clinically deployable 3D medical VLMs. Our codes will be publicly available.
LDM-Morph: Latent diffusion model guided deformable image registration
Nov 23, 2024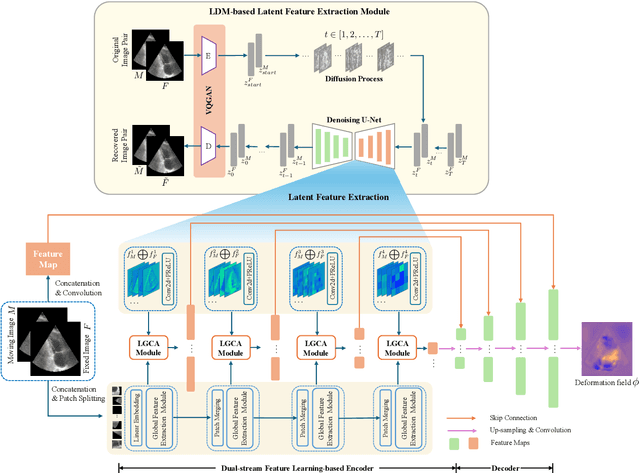
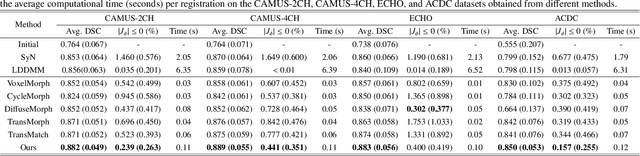
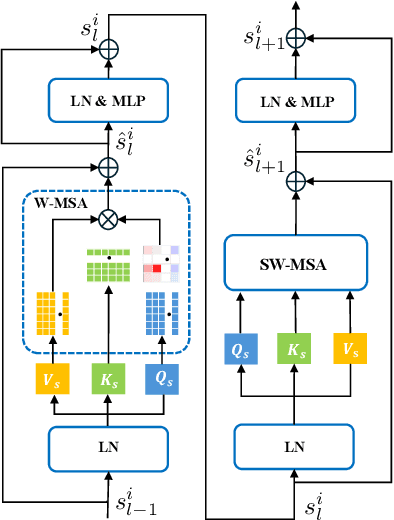
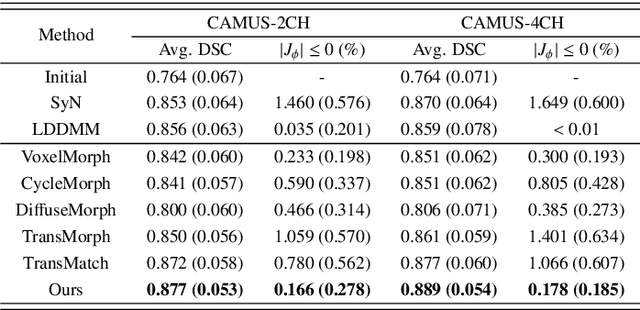
Abstract:Deformable image registration plays an essential role in various medical image tasks. Existing deep learning-based deformable registration frameworks primarily utilize convolutional neural networks (CNNs) or Transformers to learn features to predict the deformations. However, the lack of semantic information in the learned features limits the registration performance. Furthermore, the similarity metric of the loss function is often evaluated only in the pixel space, which ignores the matching of high-level anatomical features and can lead to deformation folding. To address these issues, in this work, we proposed LDM-Morph, an unsupervised deformable registration algorithm for medical image registration. LDM-Morph integrated features extracted from the latent diffusion model (LDM) to enrich the semantic information. Additionally, a latent and global feature-based cross-attention module (LGCA) was designed to enhance the interaction of semantic information from LDM and global information from multi-head self-attention operations. Finally, a hierarchical metric was proposed to evaluate the similarity of image pairs in both the original pixel space and latent-feature space, enhancing topology preservation while improving registration accuracy. Extensive experiments on four public 2D cardiac image datasets show that the proposed LDM-Morph framework outperformed existing state-of-the-art CNNs- and Transformers-based registration methods regarding accuracy and topology preservation with comparable computational efficiency. Our code is publicly available at https://github.com/wujiong-hub/LDM-Morph.
Adaptive Whole-Body PET Image Denoising Using 3D Diffusion Models with ControlNet
Nov 08, 2024


Abstract:Positron Emission Tomography (PET) is a vital imaging modality widely used in clinical diagnosis and preclinical research but faces limitations in image resolution and signal-to-noise ratio due to inherent physical degradation factors. Current deep learning-based denoising methods face challenges in adapting to the variability of clinical settings, influenced by factors such as scanner types, tracer choices, dose levels, and acquisition times. In this work, we proposed a novel 3D ControlNet-based denoising method for whole-body PET imaging. We first pre-trained a 3D Denoising Diffusion Probabilistic Model (DDPM) using a large dataset of high-quality normal-dose PET images. Following this, we fine-tuned the model on a smaller set of paired low- and normal-dose PET images, integrating low-dose inputs through a 3D ControlNet architecture, thereby making the model adaptable to denoising tasks in diverse clinical settings. Experimental results based on clinical PET datasets show that the proposed framework outperformed other state-of-the-art PET image denoising methods both in visual quality and quantitative metrics. This plug-and-play approach allows large diffusion models to be fine-tuned and adapted to PET images from diverse acquisition protocols.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge