Kuangyu Shi
Artificial intelligence for simplified patient-centered dosimetry in radiopharmaceutical therapies
Oct 14, 2025Abstract:KEY WORDS: Artificial Intelligence (AI), Theranostics, Dosimetry, Radiopharmaceutical Therapy (RPT), Patient-friendly dosimetry KEY POINTS - The rapid evolution of radiopharmaceutical therapy (RPT) highlights the growing need for personalized and patient-centered dosimetry. - Artificial Intelligence (AI) offers solutions to the key limitations in current dosimetry calculations. - The main advances on AI for simplified dosimetry toward patient-friendly RPT are reviewed. - Future directions on the role of AI in RPT dosimetry are discussed.
Fed-NDIF: A Noise-Embedded Federated Diffusion Model For Low-Count Whole-Body PET Denoising
Mar 20, 2025



Abstract:Low-count positron emission tomography (LCPET) imaging can reduce patients' exposure to radiation but often suffers from increased image noise and reduced lesion detectability, necessitating effective denoising techniques. Diffusion models have shown promise in LCPET denoising for recovering degraded image quality. However, training such models requires large and diverse datasets, which are challenging to obtain in the medical domain. To address data scarcity and privacy concerns, we combine diffusion models with federated learning -- a decentralized training approach where models are trained individually at different sites, and their parameters are aggregated on a central server over multiple iterations. The variation in scanner types and image noise levels within and across institutions poses additional challenges for federated learning in LCPET denoising. In this study, we propose a novel noise-embedded federated learning diffusion model (Fed-NDIF) to address these challenges, leveraging a multicenter dataset and varying count levels. Our approach incorporates liver normalized standard deviation (NSTD) noise embedding into a 2.5D diffusion model and utilizes the Federated Averaging (FedAvg) algorithm to aggregate locally trained models into a global model, which is subsequently fine-tuned on local datasets to optimize performance and obtain personalized models. Extensive validation on datasets from the University of Bern, Ruijin Hospital in Shanghai, and Yale-New Haven Hospital demonstrates the superior performance of our method in enhancing image quality and improving lesion quantification. The Fed-NDIF model shows significant improvements in PSNR, SSIM, and NMSE of the entire 3D volume, as well as enhanced lesion detectability and quantification, compared to local diffusion models and federated UNet-based models.
Semi-Supervised Learning for Dose Prediction in Targeted Radionuclide: A Synthetic Data Study
Mar 07, 2025Abstract:Targeted Radionuclide Therapy (TRT) is a modern strategy in radiation oncology that aims to administer a potent radiation dose specifically to cancer cells using cancer-targeting radiopharmaceuticals. Accurate radiation dose estimation tailored to individual patients is crucial. Deep learning, particularly with pre-therapy imaging, holds promise for personalizing TRT doses. However, current methods require large time series of SPECT imaging, which is hardly achievable in routine clinical practice, and thus raises issues of data availability. Our objective is to develop a semi-supervised learning (SSL) solution to personalize dosimetry using pre-therapy images. The aim is to develop an approach that achieves accurate results when PET/CT images are available, but are associated with only a few post-therapy dosimetry data provided by SPECT images. In this work, we introduce an SSL method using a pseudo-label generation approach for regression tasks inspired by the FixMatch framework. The feasibility of the proposed solution was preliminarily evaluated through an in-silico study using synthetic data and Monte Carlo simulation. Experimental results for organ dose prediction yielded promising outcomes, showing that the use of pseudo-labeled data provides better accuracy compared to using only labeled data.
Developing a PET/CT Foundation Model for Cross-Modal Anatomical and Functional Imaging
Mar 04, 2025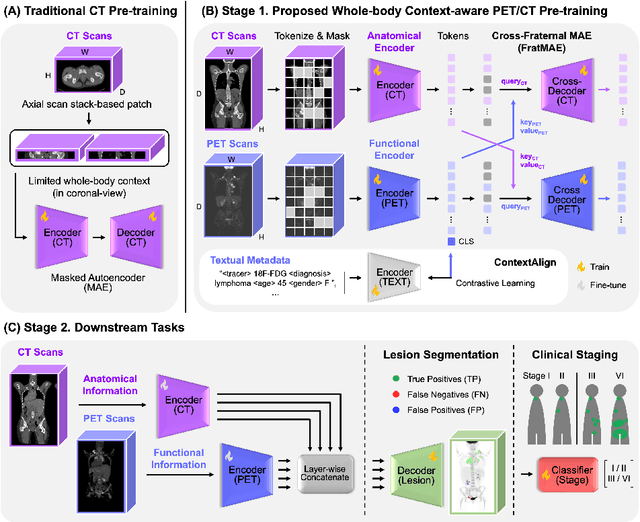
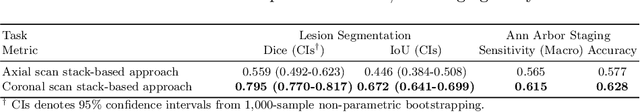
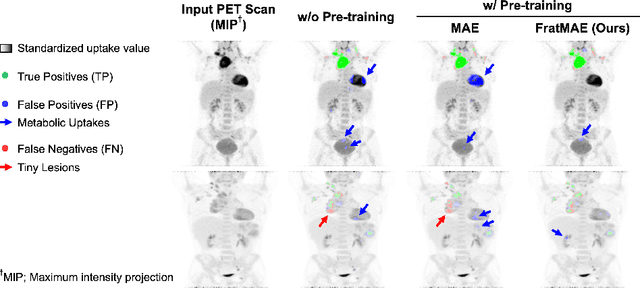
Abstract:In oncology, Positron Emission Tomography-Computed Tomography (PET/CT) is widely used in cancer diagnosis, staging, and treatment monitoring, as it combines anatomical details from CT with functional metabolic activity and molecular marker expression information from PET. However, existing artificial intelligence-driven PET/CT analyses rely predominantly on task-specific models trained from scratch or on limited datasets, limiting their generalizability and robustness. To address this, we propose a foundation model approach specifically designed for multimodal PET/CT imaging. We introduce the Cross-Fraternal Twin Masked Autoencoder (FratMAE), a novel framework that effectively integrates whole-body anatomical and functional or molecular information. FratMAE employs separate Vision Transformer (ViT) encoders for PET and CT scans, along with cross-attention decoders that enable synergistic interactions between modalities during masked autoencoder training. Additionally, it incorporates textual metadata to enhance PET representation learning. By pre-training on PET/CT datasets, FratMAE captures intricate cross-modal relationships and global uptake patterns, achieving superior performance on downstream tasks and demonstrating its potential as a generalizable foundation model.
PET Image Denoising via Text-Guided Diffusion: Integrating Anatomical Priors through Text Prompts
Feb 28, 2025Abstract:Low-dose Positron Emission Tomography (PET) imaging presents a significant challenge due to increased noise and reduced image quality, which can compromise its diagnostic accuracy and clinical utility. Denoising diffusion probabilistic models (DDPMs) have demonstrated promising performance for PET image denoising. However, existing DDPM-based methods typically overlook valuable metadata such as patient demographics, anatomical information, and scanning parameters, which should further enhance the denoising performance if considered. Recent advances in vision-language models (VLMs), particularly the pre-trained Contrastive Language-Image Pre-training (CLIP) model, have highlighted the potential of incorporating text-based information into visual tasks to improve downstream performance. In this preliminary study, we proposed a novel text-guided DDPM for PET image denoising that integrated anatomical priors through text prompts. Anatomical text descriptions were encoded using a pre-trained CLIP text encoder to extract semantic guidance, which was then incorporated into the diffusion process via the cross-attention mechanism. Evaluations based on paired 1/20 low-dose and normal-dose 18F-FDG PET datasets demonstrated that the proposed method achieved better quantitative performance than conventional UNet and standard DDPM methods at both the whole-body and organ levels. These results underscored the potential of leveraging VLMs to integrate rich metadata into the diffusion framework to enhance the image quality of low-dose PET scans.
Enhancing Global Sensitivity and Uncertainty Quantification in Medical Image Reconstruction with Monte Carlo Arbitrary-Masked Mamba
May 27, 2024Abstract:Deep learning has been extensively applied in medical image reconstruction, where Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs) represent the predominant paradigms, each possessing distinct advantages and inherent limitations: CNNs exhibit linear complexity with local sensitivity, whereas ViTs demonstrate quadratic complexity with global sensitivity. The emerging Mamba has shown superiority in learning visual representation, which combines the advantages of linear scalability and global sensitivity. In this study, we introduce MambaMIR, an Arbitrary-Masked Mamba-based model with wavelet decomposition for joint medical image reconstruction and uncertainty estimation. A novel Arbitrary Scan Masking (ASM) mechanism ``masks out'' redundant information to introduce randomness for further uncertainty estimation. Compared to the commonly used Monte Carlo (MC) dropout, our proposed MC-ASM provides an uncertainty map without the need for hyperparameter tuning and mitigates the performance drop typically observed when applying dropout to low-level tasks. For further texture preservation and better perceptual quality, we employ the wavelet transformation into MambaMIR and explore its variant based on the Generative Adversarial Network, namely MambaMIR-GAN. Comprehensive experiments have been conducted for multiple representative medical image reconstruction tasks, demonstrating that the proposed MambaMIR and MambaMIR-GAN outperform other baseline and state-of-the-art methods in different reconstruction tasks, where MambaMIR achieves the best reconstruction fidelity and MambaMIR-GAN has the best perceptual quality. In addition, our MC-ASM provides uncertainty maps as an additional tool for clinicians, while mitigating the typical performance drop caused by the commonly used dropout.
LpQcM: Adaptable Lesion-Quantification-Consistent Modulation for Deep Learning Low-Count PET Image Denoising
Apr 27, 2024Abstract:Deep learning-based positron emission tomography (PET) image denoising offers the potential to reduce radiation exposure and scanning time by transforming low-count images into high-count equivalents. However, existing methods typically blur crucial details, leading to inaccurate lesion quantification. This paper proposes a lesion-perceived and quantification-consistent modulation (LpQcM) strategy for enhanced PET image denoising, via employing downstream lesion quantification analysis as auxiliary tools. The LpQcM is a plug-and-play design adaptable to a wide range of model architectures, modulating the sampling and optimization procedures of model training without adding any computational burden to the inference phase. Specifically, the LpQcM consists of two components, the lesion-perceived modulation (LpM) and the multiscale quantification-consistent modulation (QcM). The LpM enhances lesion contrast and visibility by allocating higher sampling weights and stricter loss criteria to lesion-present samples determined by an auxiliary segmentation network than lesion-absent ones. The QcM further emphasizes accuracy of quantification for both the mean and maximum standardized uptake value (SUVmean and SUVmax) across multiscale sub-regions throughout the entire image, thereby enhancing the overall image quality. Experiments conducted on large PET datasets from multiple centers and vendors, and varying noise levels demonstrated the LpQcM efficacy across various denoising frameworks. Compared to frameworks without LpQcM, the integration of LpQcM reduces the lesion SUVmean bias by 2.92% on average and increases the peak signal-to-noise ratio (PSNR) by 0.34 on average, for denoising images of extremely low-count levels below 10%.
Self-Supervised Learning for Physiologically-Based Pharmacokinetic Modeling in Dynamic PET
May 17, 2023



Abstract:Dynamic positron emission tomography imaging (dPET) provides temporally resolved images of a tracer enabling a quantitative measure of physiological processes. Voxel-wise physiologically-based pharmacokinetic (PBPK) modeling of the time activity curves (TAC) can provide relevant diagnostic information for clinical workflow. Conventional fitting strategies for TACs are slow and ignore the spatial relation between neighboring voxels. We train a spatio-temporal UNet to estimate the kinetic parameters given TAC from F-18-fluorodeoxyglucose (FDG) dPET. This work introduces a self-supervised loss formulation to enforce the similarity between the measured TAC and those generated with the learned kinetic parameters. Our method provides quantitatively comparable results at organ-level to the significantly slower conventional approaches, while generating pixel-wise parametric images which are consistent with expected physiology. To the best of our knowledge, this is the first self-supervised network that allows voxel-wise computation of kinetic parameters consistent with a non-linear kinetic model. The code will become publicly available upon acceptance.
FedFTN: Personalized Federated Learning with Deep Feature Transformation Network for Multi-institutional Low-count PET Denoising
Apr 02, 2023Abstract:Low-count PET is an efficient way to reduce radiation exposure and acquisition time, but the reconstructed images often suffer from low signal-to-noise ratio (SNR), thus affecting diagnosis and other downstream tasks. Recent advances in deep learning have shown great potential in improving low-count PET image quality, but acquiring a large, centralized, and diverse dataset from multiple institutions for training a robust model is difficult due to privacy and security concerns of patient data. Moreover, low-count PET data at different institutions may have different data distribution, thus requiring personalized models. While previous federated learning (FL) algorithms enable multi-institution collaborative training without the need of aggregating local data, addressing the large domain shift in the application of multi-institutional low-count PET denoising remains a challenge and is still highly under-explored. In this work, we propose FedFTN, a personalized federated learning strategy that addresses these challenges. FedFTN uses a local deep feature transformation network (FTN) to modulate the feature outputs of a globally shared denoising network, enabling personalized low-count PET denoising for each institution. During the federated learning process, only the denoising network's weights are communicated and aggregated, while the FTN remains at the local institutions for feature transformation. We evaluated our method using a large-scale dataset of multi-institutional low-count PET imaging data from three medical centers located across three continents, and showed that FedFTN provides high-quality low-count PET images, outperforming previous baseline FL reconstruction methods across all low-count levels at all three institutions.
Application of the nnU-Net for automatic segmentation of lung lesion on CT images, and implication on radiomic models
Sep 24, 2022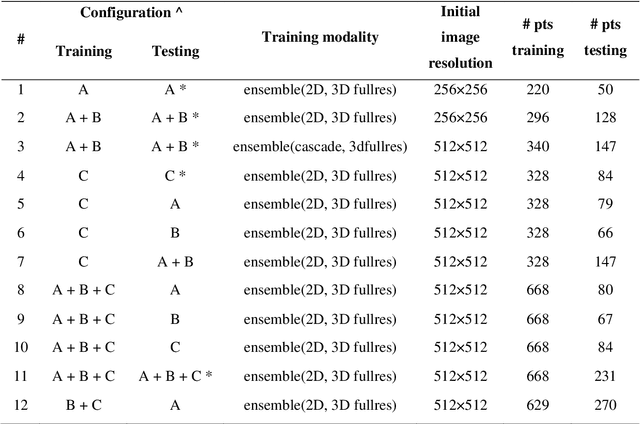
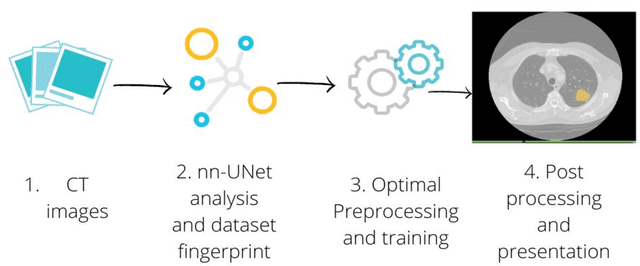
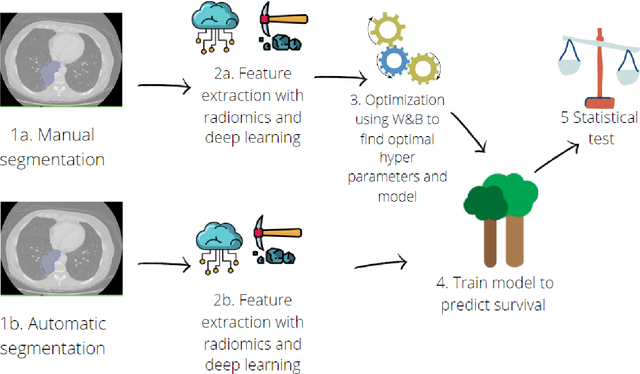
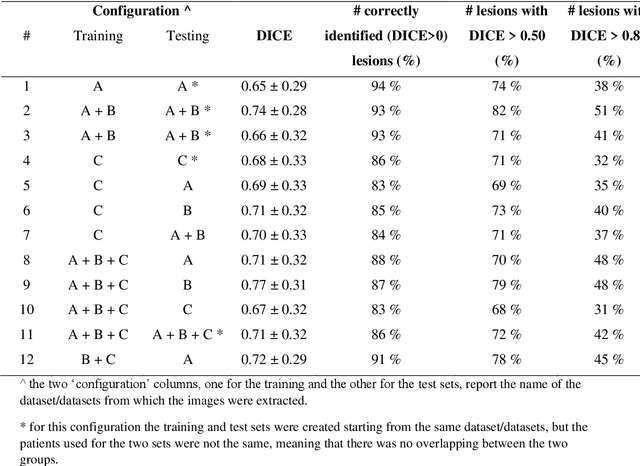
Abstract:Lesion segmentation is a crucial step of the radiomic workflow. Manual segmentation requires long execution time and is prone to variability, impairing the realisation of radiomic studies and their robustness. In this study, a deep-learning automatic segmentation method was applied on computed tomography images of non-small-cell lung cancer patients. The use of manual vs automatic segmentation in the performance of survival radiomic models was assessed, as well. METHODS A total of 899 NSCLC patients were included (2 proprietary: A and B, 1 public datasets: C). Automatic segmentation of lung lesions was performed by training a previously developed architecture, the nnU-Net, including 2D, 3D and cascade approaches. The quality of automatic segmentation was evaluated with DICE coefficient, considering manual contours as reference. The impact of automatic segmentation on the performance of a radiomic model for patient survival was explored by extracting radiomic hand-crafted and deep-learning features from manual and automatic contours of dataset A, and feeding different machine learning algorithms to classify survival above/below median. Models' accuracies were assessed and compared. RESULTS The best agreement between automatic and manual contours with DICE=0.78 +(0.12) was achieved by averaging predictions from 2D and 3D models, and applying a post-processing technique to extract the maximum connected component. No statistical differences were observed in the performances of survival models when using manual or automatic contours, hand-crafted, or deep features. The best classifier showed an accuracy between 0.65 and 0.78. CONCLUSION The promising role of nnU-Net for automatic segmentation of lung lesions was confirmed, dramatically reducing the time-consuming physicians' workload without impairing the accuracy of survival predictive models based on radiomics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge