Matteo Ferrante
Simple Models, Rich Representations: Visual Decoding from Primate Intracortical Neural Signals
Jan 16, 2026Abstract:Understanding how neural activity gives rise to perception is a central challenge in neuroscience. We address the problem of decoding visual information from high-density intracortical recordings in primates, using the THINGS Ventral Stream Spiking Dataset. We systematically evaluate the effects of model architecture, training objectives, and data scaling on decoding performance. Results show that decoding accuracy is mainly driven by modeling temporal dynamics in neural signals, rather than architectural complexity. A simple model combining temporal attention with a shallow MLP achieves up to 70% top-1 image retrieval accuracy, outperforming linear baselines as well as recurrent and convolutional approaches. Scaling analyses reveal predictable diminishing returns with increasing input dimensionality and dataset size. Building on these findings, we design a modular generative decoding pipeline that combines low-resolution latent reconstruction with semantically conditioned diffusion, generating plausible images from 200 ms of brain activity. This framework provides principles for brain-computer interfaces and semantic neural decoding.
Transforming Multimodal Models into Action Models for Radiotherapy
Feb 06, 2025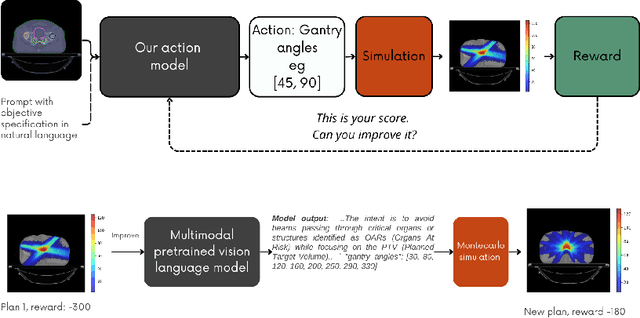
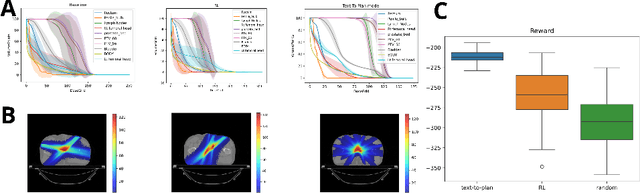
Abstract:Radiotherapy is a crucial cancer treatment that demands precise planning to balance tumor eradication and preservation of healthy tissue. Traditional treatment planning (TP) is iterative, time-consuming, and reliant on human expertise, which can potentially introduce variability and inefficiency. We propose a novel framework to transform a large multimodal foundation model (MLM) into an action model for TP using a few-shot reinforcement learning (RL) approach. Our method leverages the MLM's extensive pre-existing knowledge of physics, radiation, and anatomy, enhancing it through a few-shot learning process. This allows the model to iteratively improve treatment plans using a Monte Carlo simulator. Our results demonstrate that this method outperforms conventional RL-based approaches in both quality and efficiency, achieving higher reward scores and more optimal dose distributions in simulations on prostate cancer data. This proof-of-concept suggests a promising direction for integrating advanced AI models into clinical workflows, potentially enhancing the speed, quality, and standardization of radiotherapy treatment planning.
Towards Neural Foundation Models for Vision: Aligning EEG, MEG, and fMRI Representations for Decoding, Encoding, and Modality Conversion
Nov 14, 2024Abstract:This paper presents a novel approach towards creating a foundational model for aligning neural data and visual stimuli across multimodal representationsof brain activity by leveraging contrastive learning. We used electroencephalography (EEG), magnetoencephalography (MEG), and functional magnetic resonance imaging (fMRI) data. Our framework's capabilities are demonstrated through three key experiments: decoding visual information from neural data, encoding images into neural representations, and converting between neural modalities. The results highlight the model's ability to accurately capture semantic information across different brain imaging techniques, illustrating its potential in decoding, encoding, and modality conversion tasks.
R&B -- Rhythm and Brain: Cross-subject Decoding of Music from Human Brain Activity
Jun 21, 2024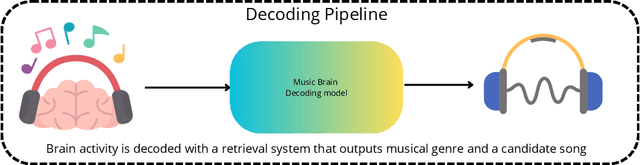

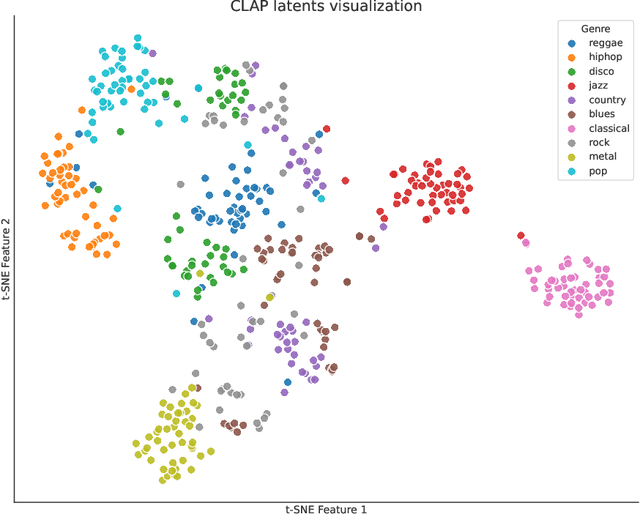
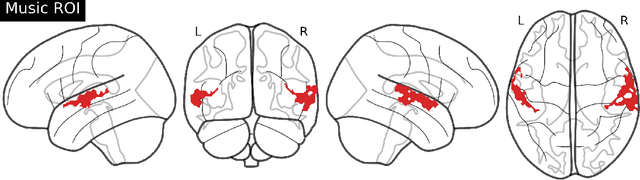
Abstract:Music is a universal phenomenon that profoundly influences human experiences across cultures. This study investigates whether music can be decoded from human brain activity measured with functional MRI (fMRI) during its perception. Leveraging recent advancements in extensive datasets and pre-trained computational models, we construct mappings between neural data and latent representations of musical stimuli. Our approach integrates functional and anatomical alignment techniques to facilitate cross-subject decoding, addressing the challenges posed by the low temporal resolution and signal-to-noise ratio (SNR) in fMRI data. Starting from the GTZan fMRI dataset, where five participants listened to 540 musical stimuli from 10 different genres while their brain activity was recorded, we used the CLAP (Contrastive Language-Audio Pretraining) model to extract latent representations of the musical stimuli and developed voxel-wise encoding models to identify brain regions responsive to these stimuli. By applying a threshold to the association between predicted and actual brain activity, we identified specific regions of interest (ROIs) which can be interpreted as key players in music processing. Our decoding pipeline, primarily retrieval-based, employs a linear map to project brain activity to the corresponding CLAP features. This enables us to predict and retrieve the musical stimuli most similar to those that originated the fMRI data. Our results demonstrate state-of-the-art identification accuracy, with our methods significantly outperforming existing approaches. Our findings suggest that neural-based music retrieval systems could enable personalized recommendations and therapeutic applications. Future work could use higher temporal resolution neuroimaging and generative models to improve decoding accuracy and explore the neural underpinnings of music perception and emotion.
Optimizing Genetically-Driven Synaptogenesis
Feb 11, 2024
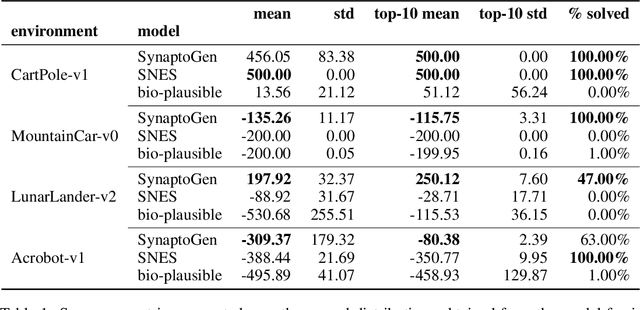

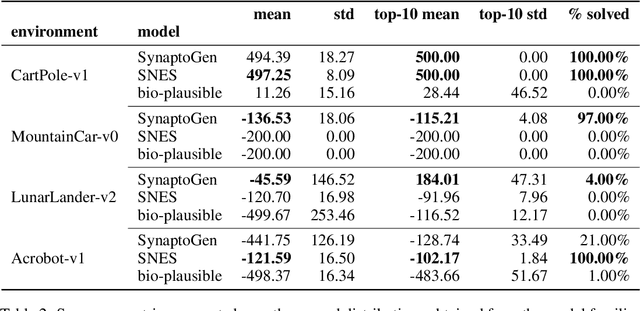
Abstract:In this paper we introduce SynaptoGen, a novel framework that aims to bridge the gap between genetic manipulations and neuronal network behavior by simulating synaptogenesis and guiding the development of neuronal networks capable of solving predetermined computational tasks. Drawing inspiration from recent advancements in the field, we propose SynaptoGen as a bio-plausible approach to modeling synaptogenesis through differentiable functions. To validate SynaptoGen, we conduct a preliminary experiment using reinforcement learning as a benchmark learning framework, demonstrating its effectiveness in generating neuronal networks capable of solving the OpenAI Gym's Cart Pole task, compared to carefully designed baselines. The results highlight the potential of SynaptoGen to inspire further advancements in neuroscience and computational modeling, while also acknowledging the need for incorporating more realistic genetic rules and synaptic conductances in future research. Overall, SynaptoGen represents a promising avenue for exploring the intersection of genetics, neuroscience, and artificial intelligence.
Decoding visual brain representations from electroencephalography through Knowledge Distillation and latent diffusion models
Sep 08, 2023Abstract:Decoding visual representations from human brain activity has emerged as a thriving research domain, particularly in the context of brain-computer interfaces. Our study presents an innovative method that employs to classify and reconstruct images from the ImageNet dataset using electroencephalography (EEG) data from subjects that had viewed the images themselves (i.e. "brain decoding"). We analyzed EEG recordings from 6 participants, each exposed to 50 images spanning 40 unique semantic categories. These EEG readings were converted into spectrograms, which were then used to train a convolutional neural network (CNN), integrated with a knowledge distillation procedure based on a pre-trained Contrastive Language-Image Pre-Training (CLIP)-based image classification teacher network. This strategy allowed our model to attain a top-5 accuracy of 80%, significantly outperforming a standard CNN and various RNN-based benchmarks. Additionally, we incorporated an image reconstruction mechanism based on pre-trained latent diffusion models, which allowed us to generate an estimate of the images which had elicited EEG activity. Therefore, our architecture not only decodes images from neural activity but also offers a credible image reconstruction from EEG only, paving the way for e.g. swift, individualized feedback experiments. Our research represents a significant step forward in connecting neural signals with visual cognition.
Brain Captioning: Decoding human brain activity into images and text
May 19, 2023
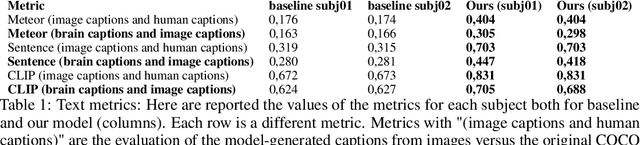

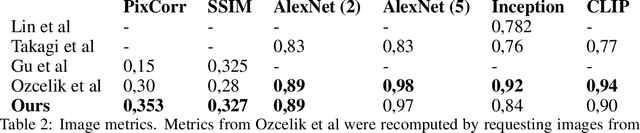
Abstract:Every day, the human brain processes an immense volume of visual information, relying on intricate neural mechanisms to perceive and interpret these stimuli. Recent breakthroughs in functional magnetic resonance imaging (fMRI) have enabled scientists to extract visual information from human brain activity patterns. In this study, we present an innovative method for decoding brain activity into meaningful images and captions, with a specific focus on brain captioning due to its enhanced flexibility as compared to brain decoding into images. Our approach takes advantage of cutting-edge image captioning models and incorporates a unique image reconstruction pipeline that utilizes latent diffusion models and depth estimation. We utilized the Natural Scenes Dataset, a comprehensive fMRI dataset from eight subjects who viewed images from the COCO dataset. We employed the Generative Image-to-text Transformer (GIT) as our backbone for captioning and propose a new image reconstruction pipeline based on latent diffusion models. The method involves training regularized linear regression models between brain activity and extracted features. Additionally, we incorporated depth maps from the ControlNet model to further guide the reconstruction process. We evaluate our methods using quantitative metrics for both generated captions and images. Our brain captioning approach outperforms existing methods, while our image reconstruction pipeline generates plausible images with improved spatial relationships. In conclusion, we demonstrate significant progress in brain decoding, showcasing the enormous potential of integrating vision and language to better understand human cognition. Our approach provides a flexible platform for future research, with potential applications in various fields, including neural art, style transfer, and portable devices.
Beyond Multilayer Perceptrons: Investigating Complex Topologies in Neural Networks
Mar 31, 2023



Abstract:In this study, we explore the impact of network topology on the approximation capabilities of artificial neural networks (ANNs), with a particular focus on complex topologies. We propose a novel methodology for constructing complex ANNs based on various topologies, including Barab\'asi-Albert, Erd\H{o}s-R\'enyi, Watts-Strogatz, and multilayer perceptrons (MLPs). The constructed networks are evaluated on synthetic datasets generated from manifold learning generators, with varying levels of task difficulty and noise. Our findings reveal that complex topologies lead to superior performance in high-difficulty regimes compared to traditional MLPs. This performance advantage is attributed to the ability of complex networks to exploit the compositionality of the underlying target function. However, this benefit comes at the cost of increased forward-pass computation time and reduced robustness to graph damage. Additionally, we investigate the relationship between various topological attributes and model performance. Our analysis shows that no single attribute can account for the observed performance differences, suggesting that the influence of network topology on approximation capabilities may be more intricate than a simple correlation with individual topological attributes. Our study sheds light on the potential of complex topologies for enhancing the performance of ANNs and provides a foundation for future research exploring the interplay between multiple topological attributes and their impact on model performance.
Semantic Brain Decoding: from fMRI to conceptually similar image reconstruction of visual stimuli
Dec 13, 2022



Abstract:Brain decoding is a field of computational neuroscience that uses measurable brain activity to infer mental states or internal representations of perceptual inputs. Therefore, we propose a novel approach to brain decoding that also relies on semantic and contextual similarity. We employ an fMRI dataset of natural image vision and create a deep learning decoding pipeline inspired by the existence of both bottom-up and top-down processes in human vision. We train a linear brain-to-feature model to map fMRI activity features to visual stimuli features, assuming that the brain projects visual information onto a space that is homeomorphic to the latent space represented by the last convolutional layer of a pretrained convolutional neural network, which typically collects a variety of semantic features that summarize and highlight similarities and differences between concepts. These features are then categorized in the latent space using a nearest-neighbor strategy, and the results are used to condition a generative latent diffusion model to create novel images. From fMRI data only, we produce reconstructions of visual stimuli that match the original content very well on a semantic level, surpassing the state of the art in previous literature. We evaluate our work and obtain good results using a quantitative semantic metric (the Wu-Palmer similarity metric over the WordNet lexicon, which had an average value of 0.57) and perform a human evaluation experiment that resulted in correct evaluation, according to the multiplicity of human criteria in evaluating image similarity, in over 80% of the test set.
BayesNetCNN: incorporating uncertainty in neural networks for image-based classification tasks
Sep 27, 2022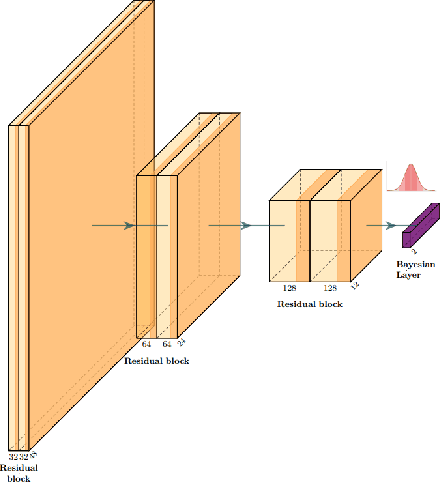
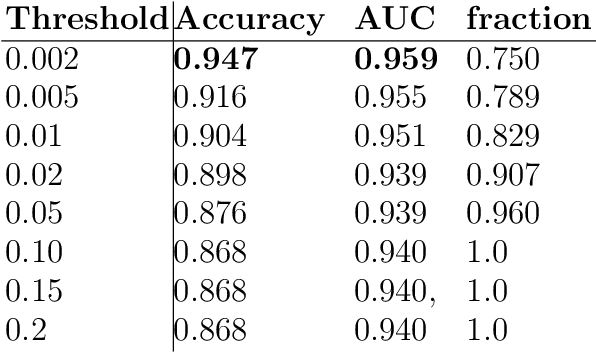
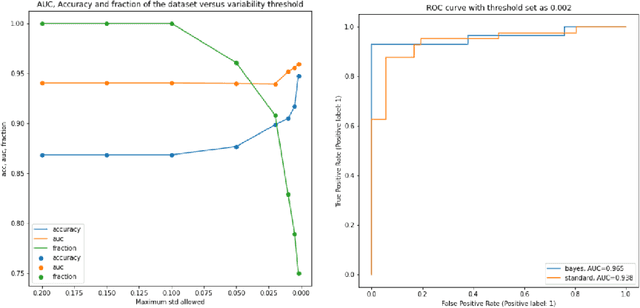
Abstract:The willingness to trust predictions formulated by automatic algorithms is key in a vast number of domains. However, a vast number of deep architectures are only able to formulate predictions without an associated uncertainty. In this paper, we propose a method to convert a standard neural network into a Bayesian neural network and estimate the variability of predictions by sampling different networks similar to the original one at each forward pass. We couple our methods with a tunable rejection-based approach that employs only the fraction of the dataset that the model is able to classify with an uncertainty below a user-set threshold. We test our model in a large cohort of brain images from Alzheimer's Disease patients, where we tackle discrimination of patients from healthy controls based on morphometric images only. We demonstrate how combining the estimated uncertainty with a rejection-based approach increases classification accuracy from 0.86 to 0.95 while retaining 75% of the test set. In addition, the model can select cases to be recommended for manual evaluation based on excessive uncertainty. We believe that being able to estimate the uncertainty of a prediction, along with tools that can modulate the behavior of the network to a degree of confidence that the user is informed about (and comfortable with) can represent a crucial step in the direction of user compliance and easier integration of deep learning tools into everyday tasks currently performed by human operators.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge