Andrea Duggento
Beyond Multilayer Perceptrons: Investigating Complex Topologies in Neural Networks
Mar 31, 2023



Abstract:In this study, we explore the impact of network topology on the approximation capabilities of artificial neural networks (ANNs), with a particular focus on complex topologies. We propose a novel methodology for constructing complex ANNs based on various topologies, including Barab\'asi-Albert, Erd\H{o}s-R\'enyi, Watts-Strogatz, and multilayer perceptrons (MLPs). The constructed networks are evaluated on synthetic datasets generated from manifold learning generators, with varying levels of task difficulty and noise. Our findings reveal that complex topologies lead to superior performance in high-difficulty regimes compared to traditional MLPs. This performance advantage is attributed to the ability of complex networks to exploit the compositionality of the underlying target function. However, this benefit comes at the cost of increased forward-pass computation time and reduced robustness to graph damage. Additionally, we investigate the relationship between various topological attributes and model performance. Our analysis shows that no single attribute can account for the observed performance differences, suggesting that the influence of network topology on approximation capabilities may be more intricate than a simple correlation with individual topological attributes. Our study sheds light on the potential of complex topologies for enhancing the performance of ANNs and provides a foundation for future research exploring the interplay between multiple topological attributes and their impact on model performance.
Multimodal and multicontrast image fusion via deep generative models
Mar 28, 2023Abstract:Recently, it has become progressively more evident that classic diagnostic labels are unable to reliably describe the complexity and variability of several clinical phenotypes. This is particularly true for a broad range of neuropsychiatric illnesses (e.g., depression, anxiety disorders, behavioral phenotypes). Patient heterogeneity can be better described by grouping individuals into novel categories based on empirically derived sections of intersecting continua that span across and beyond traditional categorical borders. In this context, neuroimaging data carry a wealth of spatiotemporally resolved information about each patient's brain. However, they are usually heavily collapsed a priori through procedures which are not learned as part of model training, and consequently not optimized for the downstream prediction task. This is because every individual participant usually comes with multiple whole-brain 3D imaging modalities often accompanied by a deep genotypic and phenotypic characterization, hence posing formidable computational challenges. In this paper we design a deep learning architecture based on generative models rooted in a modular approach and separable convolutional blocks to a) fuse multiple 3D neuroimaging modalities on a voxel-wise level, b) convert them into informative latent embeddings through heavy dimensionality reduction, c) maintain good generalizability and minimal information loss. As proof of concept, we test our architecture on the well characterized Human Connectome Project database demonstrating that our latent embeddings can be clustered into easily separable subject strata which, in turn, map to different phenotypical information which was not included in the embedding creation process. This may be of aid in predicting disease evolution as well as drug response, hence supporting mechanistic disease understanding and empowering clinical trials.
VAESim: A probabilistic approach for self-supervised prototype discovery
Sep 25, 2022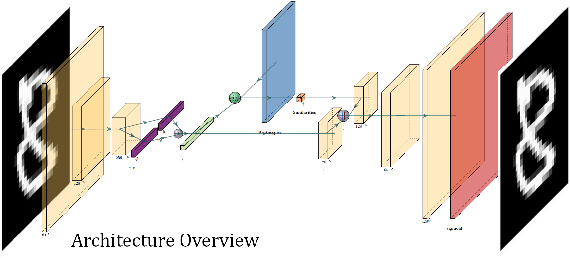
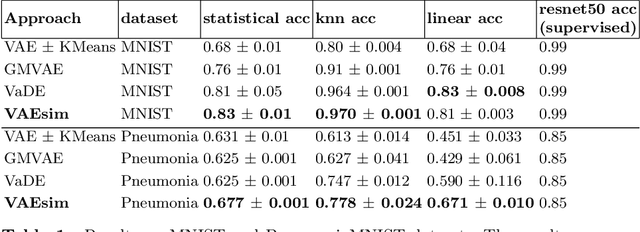
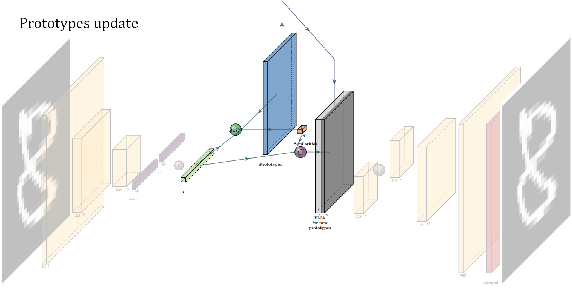
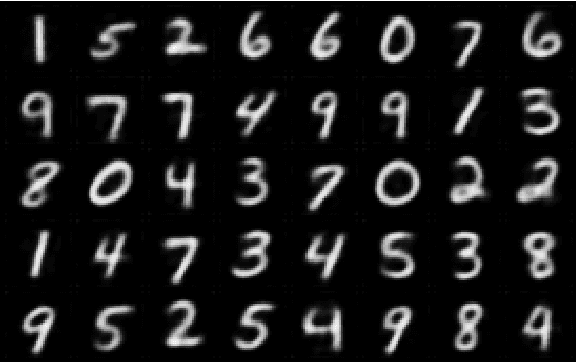
Abstract:In medicine, curated image datasets often employ discrete labels to describe what is known to be a continuous spectrum of healthy to pathological conditions, such as e.g. the Alzheimer's Disease Continuum or other areas where the image plays a pivotal point in diagnosis. We propose an architecture for image stratification based on a conditional variational autoencoder. Our framework, VAESim, leverages a continuous latent space to represent the continuum of disorders and finds clusters during training, which can then be used for image/patient stratification. The core of the method learns a set of prototypical vectors, each associated with a cluster. First, we perform a soft assignment of each data sample to the clusters. Then, we reconstruct the sample based on a similarity measure between the sample embedding and the prototypical vectors of the clusters. To update the prototypical embeddings, we use an exponential moving average of the most similar representations between actual prototypes and samples in the batch size. We test our approach on the MNIST-handwritten digit dataset and on a medical benchmark dataset called PneumoniaMNIST. We demonstrate that our method outperforms baselines in terms of kNN accuracy measured on a classification task against a standard VAE (up to 15% improvement in performance) in both datasets, and also performs at par with classification models trained in a fully supervised way. We also demonstrate how our model outperforms current, end-to-end models for unsupervised stratification.
Contrastive learning for unsupervised medical image clustering and reconstruction
Sep 24, 2022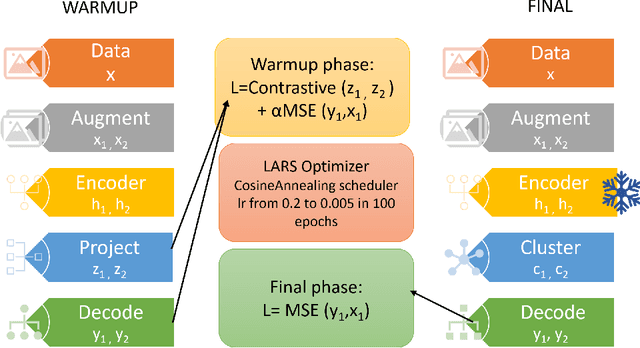
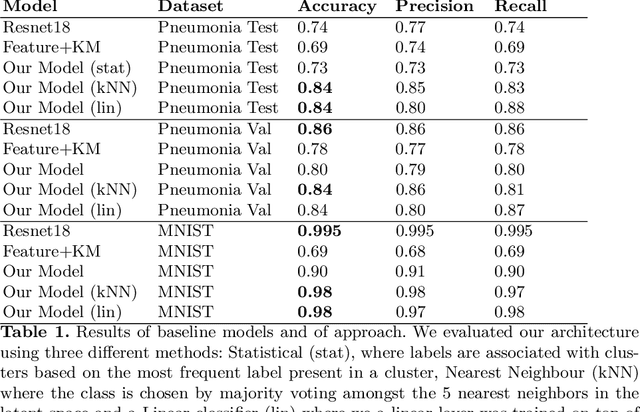
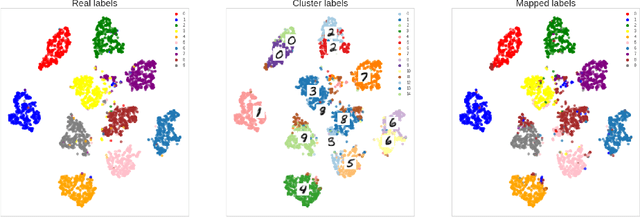
Abstract:The lack of large labeled medical imaging datasets, along with significant inter-individual variability compared to clinically established disease classes, poses significant challenges in exploiting medical imaging information in a precision medicine paradigm, where in principle dense patient-specific data can be employed to formulate individual predictions and/or stratify patients into finer-grained groups which may follow more homogeneous trajectories and therefore empower clinical trials. In order to efficiently explore the effective degrees of freedom underlying variability in medical images in an unsupervised manner, in this work we propose an unsupervised autoencoder framework which is augmented with a contrastive loss to encourage high separability in the latent space. The model is validated on (medical) benchmark datasets. As cluster labels are assigned to each example according to cluster assignments, we compare performance with a supervised transfer learning baseline. Our method achieves similar performance to the supervised architecture, indicating that separation in the latent space reproduces expert medical observer-assigned labels. The proposed method could be beneficial for patient stratification, exploring new subdivisions of larger classes or pathological continua or, due to its sampling abilities in a variation setting, data augmentation in medical image processing.
4Ward: a Relayering Strategy for Efficient Training of Arbitrarily Complex Directed Acyclic Graphs
Sep 05, 2022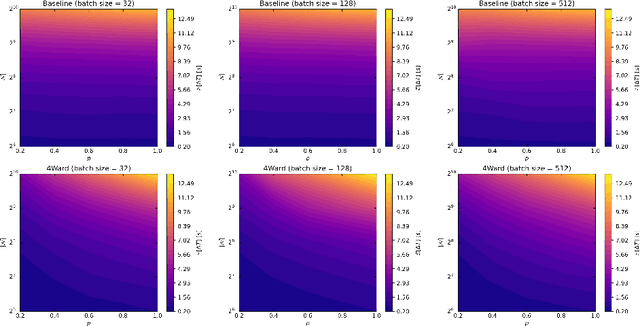
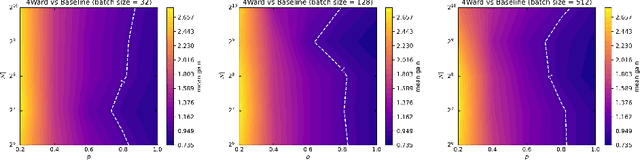
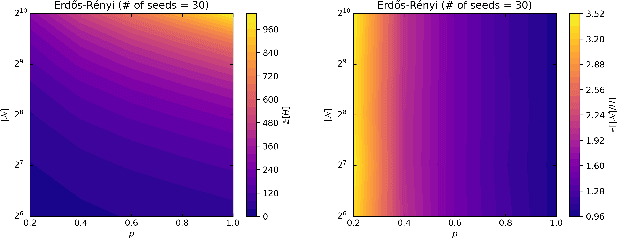
Abstract:Thanks to their ease of implementation, multilayer perceptrons (MLPs) have become ubiquitous in deep learning applications. The graph underlying an MLP is indeed multipartite, i.e. each layer of neurons only connects to neurons belonging to the adjacent layer. In constrast, in vivo brain connectomes at the level of individual synapses suggest that biological neuronal networks are characterized by scale-free degree distributions or exponentially truncated power law strength distributions, hinting at potentially novel avenues for the exploitation of evolution-derived neuronal networks. In this paper, we present "4Ward", a method and Python library capable of generating flexible and efficient neural networks (NNs) from arbitrarily complex directed acyclic graphs. 4Ward is inspired by layering algorithms drawn from the graph drawing discipline to implement efficient forward passes, and provides significant time gains in computational experiments with various Erd\H{o}s-R\'enyi graphs. 4Ward overcomes the sequential nature of the learning matrix method by parallelizing the computation of activations and provides the designer with freedom to customize weight initialization and activation functions. Our algorithm can be of aid for any investigator seeking to exploit complex topologies in a NN design framework at the microscale.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge