Giovanna Maria Dimitri
Enhancing Visual Sentiment Analysis via Semiotic Isotopy-Guided Dataset Construction
Dec 16, 2025Abstract:Visual Sentiment Analysis (VSA) is a challenging task due to the vast diversity of emotionally salient images and the inherent difficulty of acquiring sufficient data to capture this variability comprehensively. Key obstacles include building large-scale VSA datasets and developing effective methodologies that enable algorithms to identify emotionally significant elements within an image. These challenges are reflected in the limited generalization performance of VSA algorithms and models when trained and tested across different datasets. Starting from a pool of existing data collections, our approach enables the creation of a new larger dataset that not only contains a wider variety of images than the original ones, but also permits training new models with improved capability to focus on emotionally relevant combinations of image elements. This is achieved through the integration of the semiotic isotopy concept within the dataset creation process, providing deeper insights into the emotional content of images. Empirical evaluations show that models trained on a dataset generated with our method consistently outperform those trained on the original data collections, achieving superior generalization across major VSA benchmarks
Integrating Background Knowledge in Medical Semantic Segmentation with Logic Tensor Networks
Sep 26, 2025Abstract:Semantic segmentation is a fundamental task in medical image analysis, aiding medical decision-making by helping radiologists distinguish objects in an image. Research in this field has been driven by deep learning applications, which have the potential to scale these systems even in the presence of noise and artifacts. However, these systems are not yet perfected. We argue that performance can be improved by incorporating common medical knowledge into the segmentation model's loss function. To this end, we introduce Logic Tensor Networks (LTNs) to encode medical background knowledge using first-order logic (FOL) rules. The encoded rules span from constraints on the shape of the produced segmentation, to relationships between different segmented areas. We apply LTNs in an end-to-end framework with a SwinUNETR for semantic segmentation. We evaluate our method on the task of segmenting the hippocampus in brain MRI scans. Our experiments show that LTNs improve the baseline segmentation performance, especially when training data is scarce. Despite being in its preliminary stages, we argue that neurosymbolic methods are general enough to be adapted and applied to other medical semantic segmentation tasks.
A One-Class Classifier for the Detection of GAN Manipulated Multi-Spectral Satellite Images
May 19, 2023



Abstract:The highly realistic image quality achieved by current image generative models has many academic and industrial applications. To limit the use of such models to benign applications, though, it is necessary that tools to conclusively detect whether an image has been generated synthetically or not are developed. For this reason, several detectors have been developed providing excellent performance in computer vision applications, however, they can not be applied as they are to multispectral satellite images, and hence new models must be trained. In general, two-class classifiers can achieve very good detection accuracies, however they are not able to generalise to image domains and generative models architectures different than those used during training. For this reason, in this paper, we propose a one-class classifier based on Vector Quantized Variational Autoencoder 2 (VQ-VAE 2) features to overcome the limitations of two-class classifiers. First, we emphasize the generalization problem that binary classifiers suffer from by training and testing an EfficientNet-B4 architecture on multiple multispectral datasets. Then we show that, since the VQ-VAE 2 based classifier is trained only on pristine images, it is able to detect images belonging to different domains and generated by architectures that have not been used during training. Last, we compare the two classifiers head-to-head on the same generated datasets, highlighting the superiori generalization capabilities of the VQ-VAE 2-based detector.
Multimodal and multicontrast image fusion via deep generative models
Mar 28, 2023Abstract:Recently, it has become progressively more evident that classic diagnostic labels are unable to reliably describe the complexity and variability of several clinical phenotypes. This is particularly true for a broad range of neuropsychiatric illnesses (e.g., depression, anxiety disorders, behavioral phenotypes). Patient heterogeneity can be better described by grouping individuals into novel categories based on empirically derived sections of intersecting continua that span across and beyond traditional categorical borders. In this context, neuroimaging data carry a wealth of spatiotemporally resolved information about each patient's brain. However, they are usually heavily collapsed a priori through procedures which are not learned as part of model training, and consequently not optimized for the downstream prediction task. This is because every individual participant usually comes with multiple whole-brain 3D imaging modalities often accompanied by a deep genotypic and phenotypic characterization, hence posing formidable computational challenges. In this paper we design a deep learning architecture based on generative models rooted in a modular approach and separable convolutional blocks to a) fuse multiple 3D neuroimaging modalities on a voxel-wise level, b) convert them into informative latent embeddings through heavy dimensionality reduction, c) maintain good generalizability and minimal information loss. As proof of concept, we test our architecture on the well characterized Human Connectome Project database demonstrating that our latent embeddings can be clustered into easily separable subject strata which, in turn, map to different phenotypical information which was not included in the embedding creation process. This may be of aid in predicting disease evolution as well as drug response, hence supporting mechanistic disease understanding and empowering clinical trials.
A Multi-Modal Machine Learning Approach to Detect Extreme Rainfall Events in Sicily
Dec 14, 2022Abstract:In 2021 300 mm of rain, nearly half the average annual rainfall, fell near Catania (Sicily island, Italy). Such events took place in just a few hours, with dramatic consequences on the environmental, social, economic, and health systems of the region. This is the reason why, detecting extreme rainfall events is a crucial prerequisite for planning actions able to reverse possibly intensified dramatic future scenarios. In this paper, the Affinity Propagation algorithm, a clustering algorithm grounded on machine learning, was applied, to the best of our knowledge, for the first time, to identify excess rain events in Sicily. This was possible by using a high-frequency, large dataset we collected, ranging from 2009 to 2021 which we named RSE (the Rainfall Sicily Extreme dataset). Weather indicators were then been employed to validate the results, thus confirming the presence of recent anomalous rainfall events in eastern Sicily. We believe that easy-to-use and multi-modal data science techniques, such as the one proposed in this study, could give rise to significant improvements in policy-making for successfully contrasting climate changes.
Which country is this picture from? New data and methods for DNN-based country recognition
Sep 02, 2022
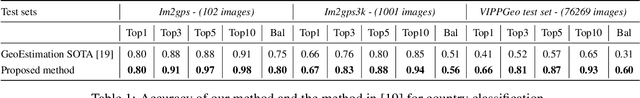


Abstract:Predicting the country where a picture has been taken from has many potential applications, like detection of false claims, impostors identification, prevention of disinformation campaigns, identification of fake news and so on. Previous works have focused mostly on the estimation of the geo-coordinates where a picture has been taken. Yet, recognizing the country where an image has been taken could potentially be more important, from a semantic and forensic point of view, than identifying its spatial coordinates. So far only a few works have addressed this task, mostly by relying on images containing characteristic landmarks, like iconic monuments. In the above framework, this paper provides two main contributions. First, we introduce a new dataset, the VIPPGeo dataset, containing almost 4 million images, that can be used to train DL models for country classification. The dataset contains only urban images given the relevance of this kind of image for country recognition, and it has been built by paying attention to removing non-significant images, like images portraying faces or specific, non-relevant objects, like airplanes or ships. Secondly, we used the dataset to train a deep learning architecture casting the country recognition problem as a classification problem. The experiments, we performed, show that our network provides significantly better results than current state of the art. In particular, we found that asking the network to directly identify the country provides better results than estimating the geo-coordinates first and then using them to trace back to the country where the picture was taken.
Modular multi-source prediction of drug side-effects with DruGNN
Feb 15, 2022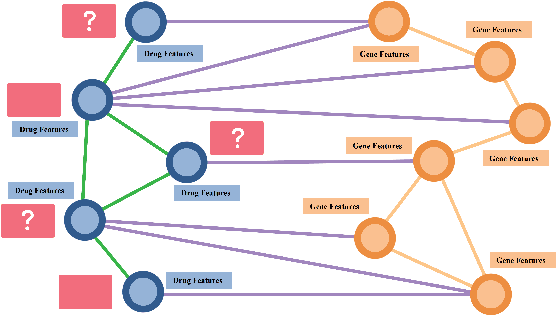
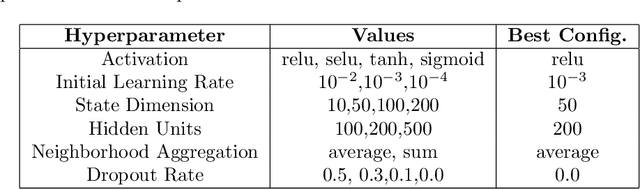
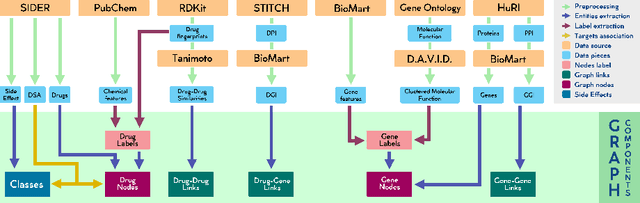
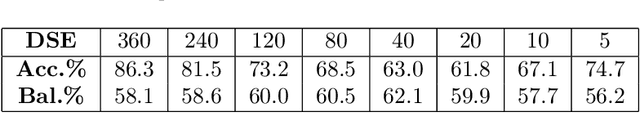
Abstract:Drug Side-Effects (DSEs) have a high impact on public health, care system costs, and drug discovery processes. Predicting the probability of side-effects, before their occurrence, is fundamental to reduce this impact, in particular on drug discovery. Candidate molecules could be screened before undergoing clinical trials, reducing the costs in time, money, and health of the participants. Drug side-effects are triggered by complex biological processes involving many different entities, from drug structures to protein-protein interactions. To predict their occurrence, it is necessary to integrate data from heterogeneous sources. In this work, such heterogeneous data is integrated into a graph dataset, expressively representing the relational information between different entities, such as drug molecules and genes. The relational nature of the dataset represents an important novelty for drug side-effect predictors. Graph Neural Networks (GNNs) are exploited to predict DSEs on our dataset with very promising results. GNNs are deep learning models that can process graph-structured data, with minimal information loss, and have been applied on a wide variety of biological tasks. Our experimental results confirm the advantage of using relationships between data entities, suggesting interesting future developments in this scope. The experimentation also shows the importance of specific subsets of data in determining associations between drugs and side-effects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge