Pietro Bongini
WILD: a new in-the-Wild Image Linkage Dataset for synthetic image attribution
Apr 29, 2025Abstract:Synthetic image source attribution is an open challenge, with an increasing number of image generators being released yearly. The complexity and the sheer number of available generative techniques, as well as the scarcity of high-quality open source datasets of diverse nature for this task, make training and benchmarking synthetic image source attribution models very challenging. WILD is a new in-the-Wild Image Linkage Dataset designed to provide a powerful training and benchmarking tool for synthetic image attribution models. The dataset is built out of a closed set of 10 popular commercial generators, which constitutes the training base of attribution models, and an open set of 10 additional generators, simulating a real-world in-the-wild scenario. Each generator is represented by 1,000 images, for a total of 10,000 images in the closed set and 10,000 images in the open set. Half of the images are post-processed with a wide range of operators. WILD allows benchmarking attribution models in a wide range of tasks, including closed and open set identification and verification, and robust attribution with respect to post-processing and adversarial attacks. Models trained on WILD are expected to benefit from the challenging scenario represented by the dataset itself. Moreover, an assessment of seven baseline methodologies on closed and open set attribution is presented, including robustness tests with respect to post-processing.
A Deep Learning Approach to the Prediction of Drug Side-Effects on Molecular Graphs
Nov 30, 2022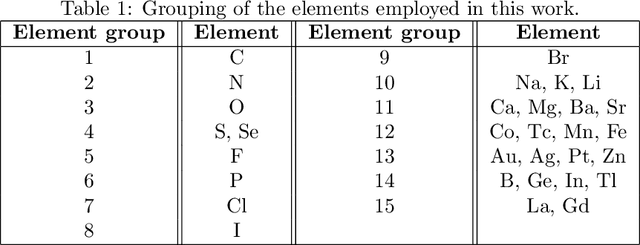
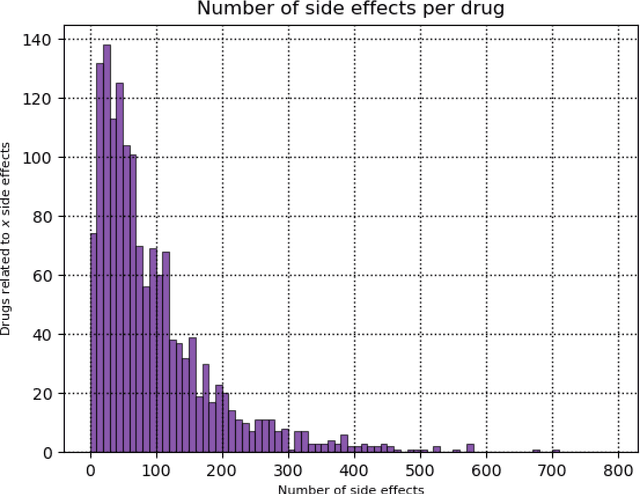
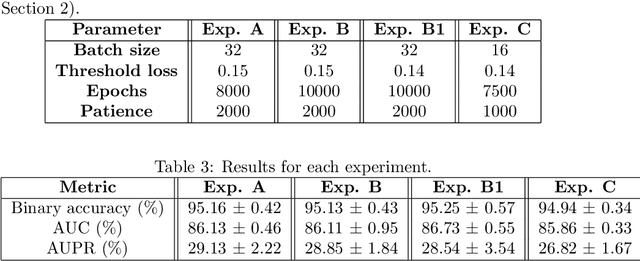
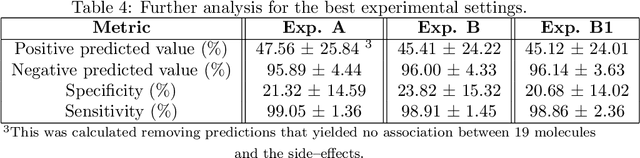
Abstract:Predicting drug side-effects before they occur is a key task in keeping the number of drug-related hospitalizations low and to improve drug discovery processes. Automatic predictors of side-effects generally are not able to process the structure of the drug, resulting in a loss of information. Graph neural networks have seen great success in recent years, thanks to their ability of exploiting the information conveyed by the graph structure and labels. These models have been used in a wide variety of biological applications, among which the prediction of drug side-effects on a large knowledge graph. Exploiting the molecular graph encoding the structure of the drug represents a novel approach, in which the problem is formulated as a multi-class multi-label graph-focused classification. We developed a methodology to carry out this task, using recurrent Graph Neural Networks, and building a dataset from freely accessible and well established data sources. The results show that our method has an improved classification capability, under many parameters and metrics, with respect to previously available predictors.
Is GPT-3 all you need for Visual Question Answering in Cultural Heritage?
Jul 25, 2022
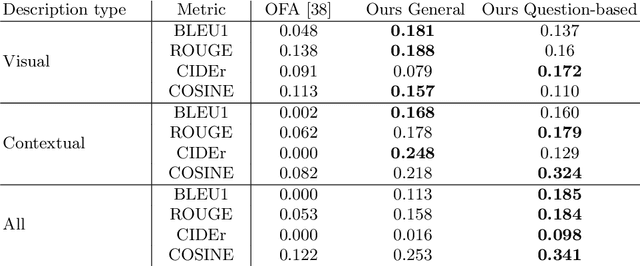

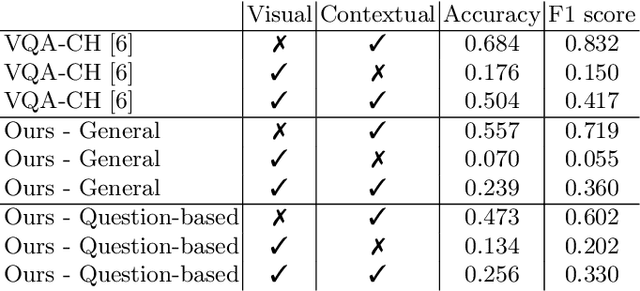
Abstract:The use of Deep Learning and Computer Vision in the Cultural Heritage domain is becoming highly relevant in the last few years with lots of applications about audio smart guides, interactive museums and augmented reality. All these technologies require lots of data to work effectively and be useful for the user. In the context of artworks, such data is annotated by experts in an expensive and time consuming process. In particular, for each artwork, an image of the artwork and a description sheet have to be collected in order to perform common tasks like Visual Question Answering. In this paper we propose a method for Visual Question Answering that allows to generate at runtime a description sheet that can be used for answering both visual and contextual questions about the artwork, avoiding completely the image and the annotation process. For this purpose, we investigate on the use of GPT-3 for generating descriptions for artworks analyzing the quality of generated descriptions through captioning metrics. Finally we evaluate the performance for Visual Question Answering and captioning tasks.
Modular multi-source prediction of drug side-effects with DruGNN
Feb 15, 2022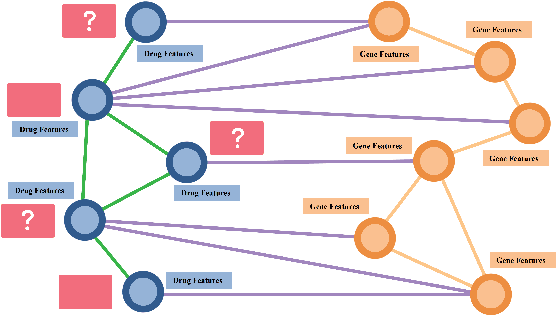
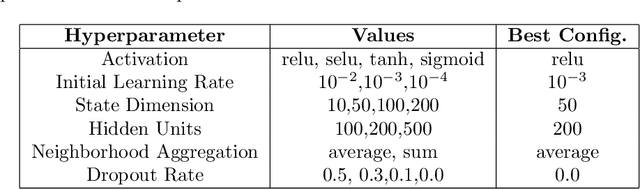
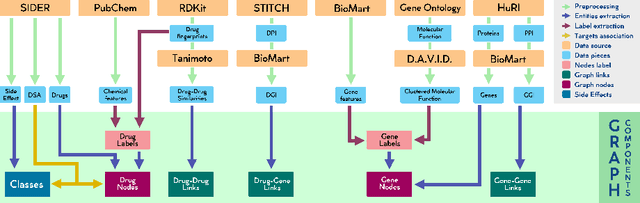
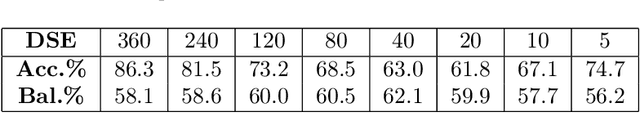
Abstract:Drug Side-Effects (DSEs) have a high impact on public health, care system costs, and drug discovery processes. Predicting the probability of side-effects, before their occurrence, is fundamental to reduce this impact, in particular on drug discovery. Candidate molecules could be screened before undergoing clinical trials, reducing the costs in time, money, and health of the participants. Drug side-effects are triggered by complex biological processes involving many different entities, from drug structures to protein-protein interactions. To predict their occurrence, it is necessary to integrate data from heterogeneous sources. In this work, such heterogeneous data is integrated into a graph dataset, expressively representing the relational information between different entities, such as drug molecules and genes. The relational nature of the dataset represents an important novelty for drug side-effect predictors. Graph Neural Networks (GNNs) are exploited to predict DSEs on our dataset with very promising results. GNNs are deep learning models that can process graph-structured data, with minimal information loss, and have been applied on a wide variety of biological tasks. Our experimental results confirm the advantage of using relationships between data entities, suggesting interesting future developments in this scope. The experimentation also shows the importance of specific subsets of data in determining associations between drugs and side-effects.
Molecular graph generation with Graph Neural Networks
Dec 14, 2020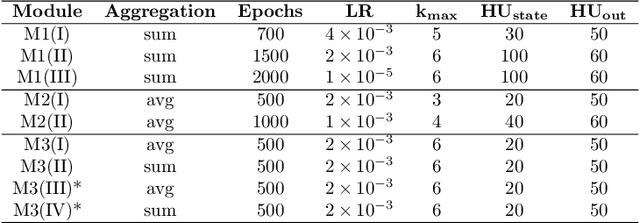
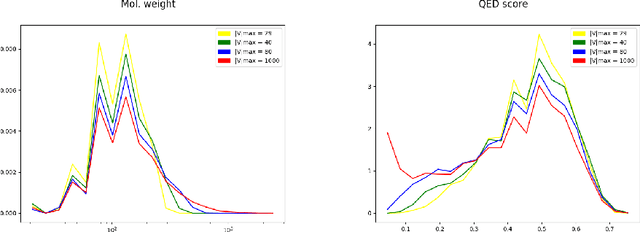
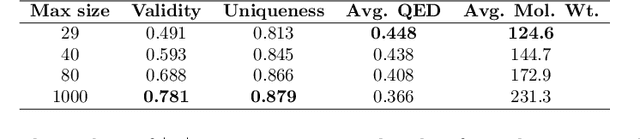
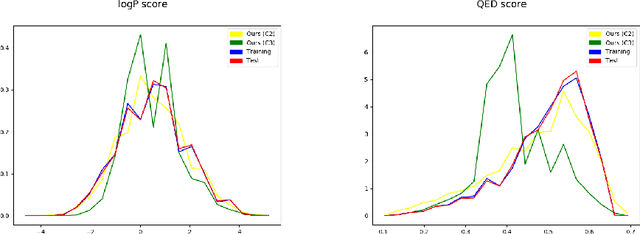
Abstract:The generation of graph-structured data is an emerging problem in the field of deep learning. Various solutions have been proposed in the last few years, yet the exploration of this branch is still in an early phase. In sequential approaches, the construction of a graph is the result of a sequence of decisions, in which, at each step, a node or a group of nodes is added to the graph, along with its connections. A very relevant application of graph generation methods is the discovery of new drug molecules, which are naturally represented as graphs. In this paper, we introduce a sequential molecular graph generator based on a set of graph neural network modules, which we call MG^2N^2. Its modular architecture simplifies the training procedure, also allowing an independent retraining of a single module. The use of graph neural networks maximizes the information in input at each generative step, which consists of the subgraph produced during the previous steps. Experiments of unconditional generation on the QM9 dataset show that our model is capable of generalizing molecular patterns seen during the training phase, without overfitting. The results indicate that our method outperforms very competitive baselines, and can be placed among the state of the art approaches for unconditional generation on QM9.
Visual Question Answering for Cultural Heritage
Mar 22, 2020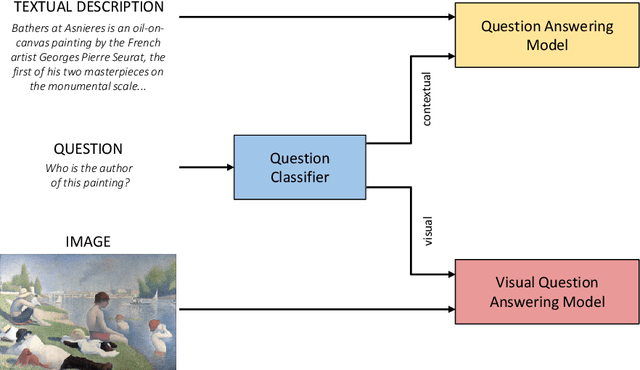
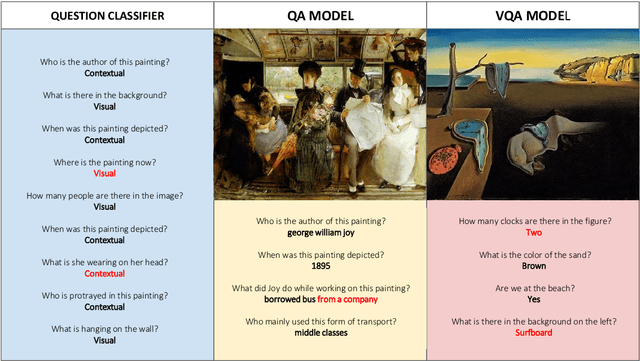
Abstract:Technology and the fruition of cultural heritage are becoming increasingly more entwined, especially with the advent of smart audio guides, virtual and augmented reality, and interactive installations. Machine learning and computer vision are important components of this ongoing integration, enabling new interaction modalities between user and museum. Nonetheless, the most frequent way of interacting with paintings and statues still remains taking pictures. Yet images alone can only convey the aesthetics of the artwork, lacking is information which is often required to fully understand and appreciate it. Usually this additional knowledge comes both from the artwork itself (and therefore the image depicting it) and from an external source of knowledge, such as an information sheet. While the former can be inferred by computer vision algorithms, the latter needs more structured data to pair visual content with relevant information. Regardless of its source, this information still must be be effectively transmitted to the user. A popular emerging trend in computer vision is Visual Question Answering (VQA), in which users can interact with a neural network by posing questions in natural language and receiving answers about the visual content. We believe that this will be the evolution of smart audio guides for museum visits and simple image browsing on personal smartphones. This will turn the classic audio guide into a smart personal instructor with which the visitor can interact by asking for explanations focused on specific interests. The advantages are twofold: on the one hand the cognitive burden of the visitor will decrease, limiting the flow of information to what the user actually wants to hear; and on the other hand it proposes the most natural way of interacting with a guide, favoring engagement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge