VAESim: A probabilistic approach for self-supervised prototype discovery
Paper and Code
Sep 25, 2022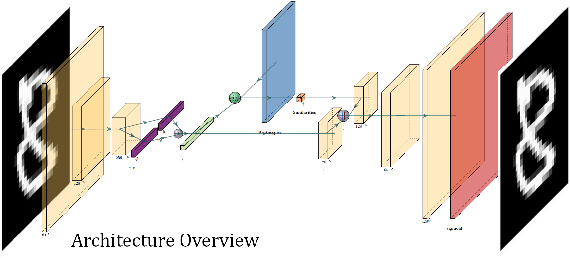
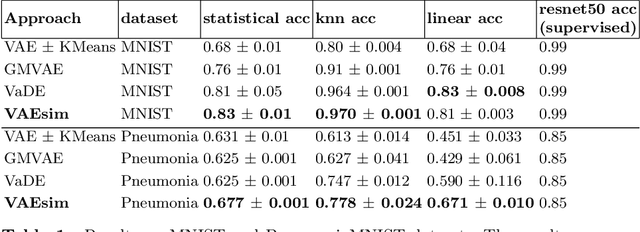
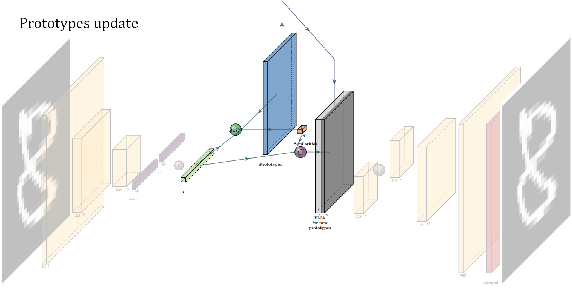
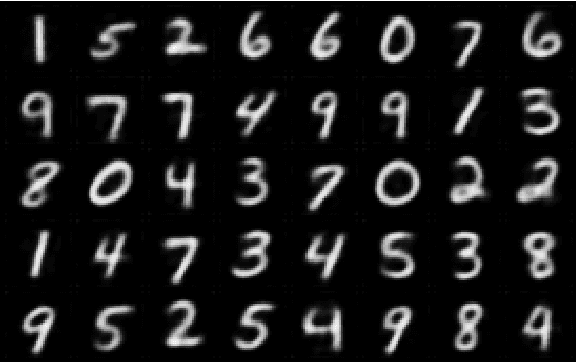
In medicine, curated image datasets often employ discrete labels to describe what is known to be a continuous spectrum of healthy to pathological conditions, such as e.g. the Alzheimer's Disease Continuum or other areas where the image plays a pivotal point in diagnosis. We propose an architecture for image stratification based on a conditional variational autoencoder. Our framework, VAESim, leverages a continuous latent space to represent the continuum of disorders and finds clusters during training, which can then be used for image/patient stratification. The core of the method learns a set of prototypical vectors, each associated with a cluster. First, we perform a soft assignment of each data sample to the clusters. Then, we reconstruct the sample based on a similarity measure between the sample embedding and the prototypical vectors of the clusters. To update the prototypical embeddings, we use an exponential moving average of the most similar representations between actual prototypes and samples in the batch size. We test our approach on the MNIST-handwritten digit dataset and on a medical benchmark dataset called PneumoniaMNIST. We demonstrate that our method outperforms baselines in terms of kNN accuracy measured on a classification task against a standard VAE (up to 15% improvement in performance) in both datasets, and also performs at par with classification models trained in a fully supervised way. We also demonstrate how our model outperforms current, end-to-end models for unsupervised stratification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge