Sailesh Conjeti
FastSurfer -- A fast and accurate deep learning based neuroimaging pipeline
Oct 09, 2019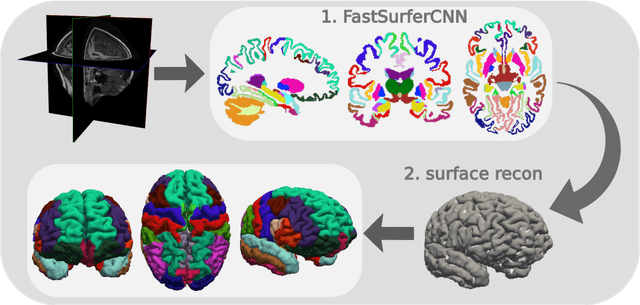
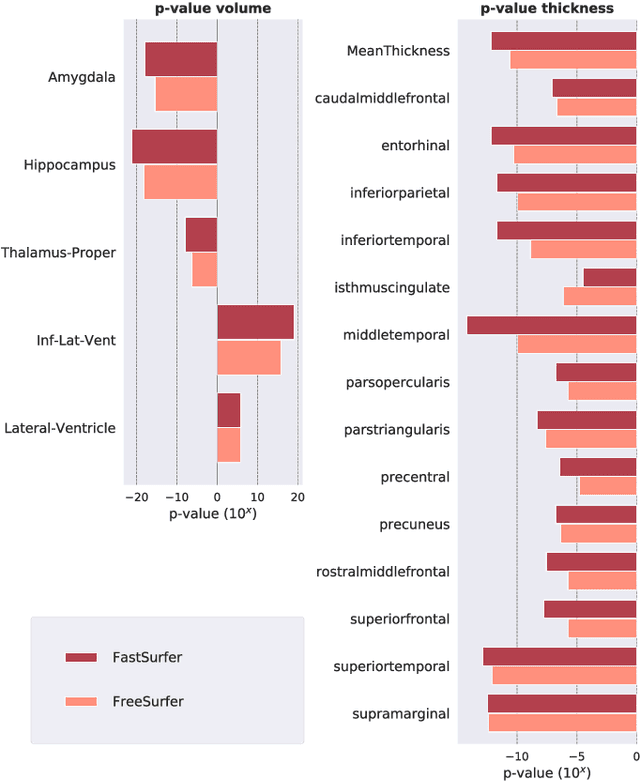
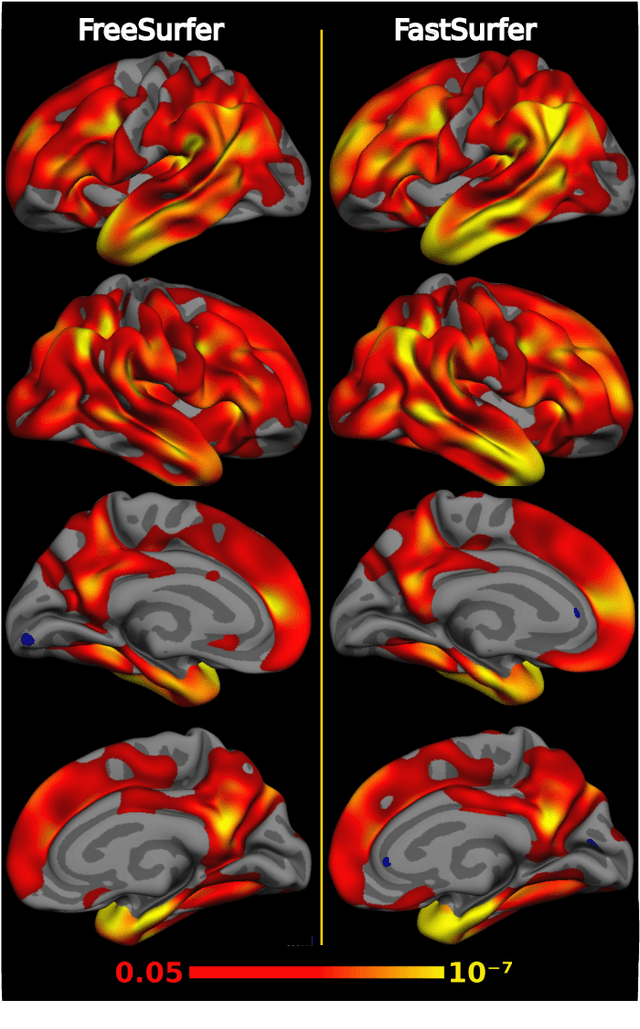
Abstract:Traditional neuroimage analysis pipelines involve computationally intensive, time-consuming optimization steps, and thus, do not scale well to large cohort studies with thousands or tens of thousands of individuals. In this work we propose a fast and accurate deep learning based neuroimaging pipeline for the automated processing of structural human brain MRI scans, including surface reconstruction and cortical parcellation. To this end, we introduce an advanced deep learning architecture capable of whole brain segmentation into 95 classes in under 1 minute, mimicking FreeSurfer's anatomical segmentation and cortical parcellation. The network architecture incorporates local and global competition via competitive dense blocks and competitive skip pathways, as well as multi-slice information aggregation that specifically tailor network performance towards accurate segmentation of both cortical and sub-cortical structures. Further, we perform fast cortical surface reconstruction and thickness analysis by introducing a spectral spherical embedding and by directly mapping the cortical labels from the image to the surface. This approach provides a full FreeSurfer alternative for volumetric analysis (within 1 minute) and surface-based thickness analysis (within only around 1h run time). For sustainability of this approach we perform extensive validation: we assert high segmentation accuracy on several unseen datasets, measure generalizability and demonstrate increased test-retest reliability, and increased sensitivity to disease effects relative to traditional FreeSurfer.
FatSegNet : A Fully Automated Deep Learning Pipeline for Adipose Tissue Segmentation on Abdominal Dixon MRI
Apr 03, 2019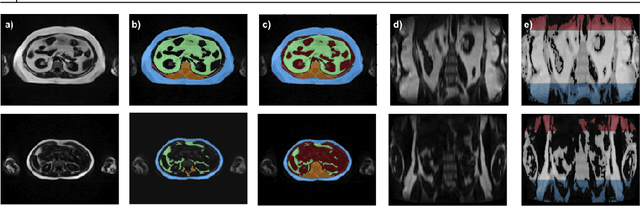



Abstract:Purpose: Development of a fast and fully automated deep learning pipeline (FatSegNet) to accurately identify, segment, and quantify abdominal adipose tissue on Dixon MRI from the Rhineland Study - a large prospective population-based study. Method: FatSegNet is composed of three stages: (i) consistent localization of the abdominal region using two 2D-Competitive Dense Fully Convolutional Networks (CDFNet), (ii) segmentation of adipose tissue on three views by independent CDFNets, and (iii) view aggregation. FatSegNet is trained with 33 manually annotated subjects, and validated by: 1) comparison of segmentation accuracy against a testingset covering a wide range of body mass index (BMI), 2) test-retest reliability, and 3) robustness in a large cohort study. Results: The CDFNet demonstrates increased robustness compared to traditional deep learning networks. FatSegNet dice score outperforms manual raters on the abdominal visceral adipose tissue (VAT, 0.828 vs. 0.788), and produces comparable results on subcutaneous adipose tissue (SAT, 0.973 vs. 0.982). The pipeline has very small test-retest absolute percentage difference and excellent agreement between scan sessions (VAT: APD = 2.957%, ICC=0.998 and SAT: APD= 3.254%, ICC=0.996). Conclusion: FatSegNet can reliably analyze a 3D Dixon MRI in1 min. It generalizes well to different body shapes, sensitively replicates known VAT and SAT volume effects in a large cohort study, and permits localized analysis of fat compartments.
Bayesian QuickNAT: Model Uncertainty in Deep Whole-Brain Segmentation for Structure-wise Quality Control
Nov 24, 2018



Abstract:We introduce Bayesian QuickNAT for the automated quality control of whole-brain segmentation on MRI T1 scans. Next to the Bayesian fully convolutional neural network, we also present inherent measures of segmentation uncertainty that allow for quality control per brain structure. For estimating model uncertainty, we follow a Bayesian approach, wherein, Monte Carlo (MC) samples from the posterior distribution are generated by keeping the dropout layers active at test time. Entropy over the MC samples provides a voxel-wise model uncertainty map, whereas expectation over the MC predictions provides the final segmentation. Next to voxel-wise uncertainty, we introduce four metrics to quantify structure-wise uncertainty in segmentation for quality control. We report experiments on four out-of-sample datasets comprising of diverse age range, pathology and imaging artifacts. The proposed structure-wise uncertainty metrics are highly correlated with the Dice score estimated with manual annotation and therefore present an inherent measure of segmentation quality. In particular, the intersection over union over all the MC samples is a suitable proxy for the Dice score. In addition to quality control at scan-level, we propose to incorporate the structure-wise uncertainty as a measure of confidence to do reliable group analysis on large data repositories. We envisage that the introduced uncertainty metrics would help assess the fidelity of automated deep learning based segmentation methods for large-scale population studies, as they enable automated quality control and group analyses in processing large data repositories.
InfiNet: Fully Convolutional Networks for Infant Brain MRI Segmentation
Oct 11, 2018



Abstract:We present a novel, parameter-efficient and practical fully convolutional neural network architecture, termed InfiNet, aimed at voxel-wise semantic segmentation of infant brain MRI images at iso-intense stage, which can be easily extended for other segmentation tasks involving multi-modalities. InfiNet consists of double encoder arms for T1 and T2 input scans that feed into a joint-decoder arm that terminates in the classification layer. The novelty of InfiNet lies in the manner in which the decoder upsamples lower resolution input feature map(s) from multiple encoder arms. Specifically, the pooled indices computed in the max-pooling layers of each of the encoder blocks are related to the corresponding decoder block to perform non-linear learning-free upsampling. The sparse maps are concatenated with intermediate encoder representations (skip connections) and convolved with trainable filters to produce dense feature maps. InfiNet is trained end-to-end to optimize for the Generalized Dice Loss, which is well-suited for high class imbalance. InfiNet achieves the whole-volume segmentation in under 50 seconds and we demonstrate competitive performance against multiple state-of-the art deep architectures and their multi-modal variants.
* 4 pages, 3 figures, conference, IEEE ISBI, 2018
Learning Optimal Deep Projection of $^{18}$F-FDG PET Imaging for Early Differential Diagnosis of Parkinsonian Syndromes
Oct 11, 2018


Abstract:Several diseases of parkinsonian syndromes present similar symptoms at early stage and no objective widely used diagnostic methods have been approved until now. Positron emission tomography (PET) with $^{18}$F-FDG was shown to be able to assess early neuronal dysfunction of synucleinopathies and tauopathies. Tensor factorization (TF) based approaches have been applied to identify characteristic metabolic patterns for differential diagnosis. However, these conventional dimension-reduction strategies assume linear or multi-linear relationships inside data, and are therefore insufficient to distinguish nonlinear metabolic differences between various parkinsonian syndromes. In this paper, we propose a Deep Projection Neural Network (DPNN) to identify characteristic metabolic pattern for early differential diagnosis of parkinsonian syndromes. We draw our inspiration from the existing TF methods. The network consists of a (i) compression part: which uses a deep network to learn optimal 2D projections of 3D scans, and a (ii) classification part: which maps the 2D projections to labels. The compression part can be pre-trained using surplus unlabelled datasets. Also, as the classification part operates on these 2D projections, it can be trained end-to-end effectively with limited labelled data, in contrast to 3D approaches. We show that DPNN is more effective in comparison to existing state-of-the-art and plausible baselines.
* 8 pages, 3 figures, conference, MICCAI DLMIA, 2018
Human Motion Analysis with Deep Metric Learning
Aug 06, 2018



Abstract:Effectively measuring the similarity between two human motions is necessary for several computer vision tasks such as gait analysis, person identi- fication and action retrieval. Nevertheless, we believe that traditional approaches such as L2 distance or Dynamic Time Warping based on hand-crafted local pose metrics fail to appropriately capture the semantic relationship across motions and, as such, are not suitable for being employed as metrics within these tasks. This work addresses this limitation by means of a triplet-based deep metric learning specifically tailored to deal with human motion data, in particular with the prob- lem of varying input size and computationally expensive hard negative mining due to motion pair alignment. Specifically, we propose (1) a novel metric learn- ing objective based on a triplet architecture and Maximum Mean Discrepancy; as well as, (2) a novel deep architecture based on attentive recurrent neural networks. One benefit of our objective function is that it enforces a better separation within the learned embedding space of the different motion categories by means of the associated distribution moments. At the same time, our attentive recurrent neural network allows processing varying input sizes to a fixed size of embedding while learning to focus on those motion parts that are semantically distinctive. Our ex- periments on two different datasets demonstrate significant improvements over conventional human motion metrics.
Competition vs. Concatenation in Skip Connections of Fully Convolutional Networks
Jul 20, 2018



Abstract:Increased information sharing through short and long-range skip connections between layers in fully convolutional networks have demonstrated significant improvement in performance for semantic segmentation. In this paper, we propose Competitive Dense Fully Convolutional Networks (CDFNet) by introducing competitive maxout activations in place of naive feature concatenation for inducing competition amongst layers. Within CDFNet, we propose two architectural contributions, namely competitive dense block (CDB) and competitive unpooling block (CUB) to induce competition at local and global scales for short and long-range skip connections respectively. This extension is demonstrated to boost learning of specialized sub-networks targeted at segmenting specific anatomies, which in turn eases the training of complex tasks. We present the proof-of-concept on the challenging task of whole body segmentation in the publicly available VISCERAL benchmark and demonstrate improved performance over multiple learning and registration based state-of-the-art methods.
Complex Fully Convolutional Neural Networks for MR Image Reconstruction
Jul 09, 2018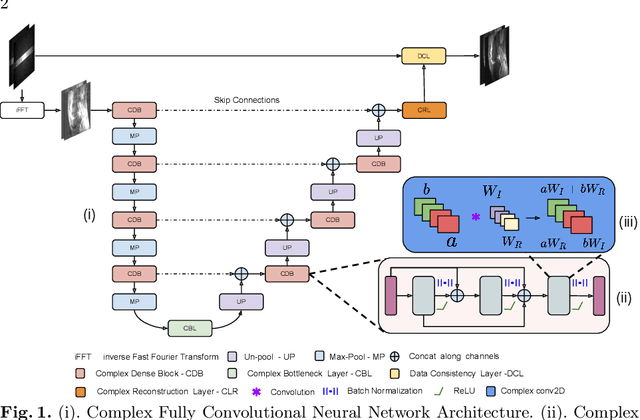
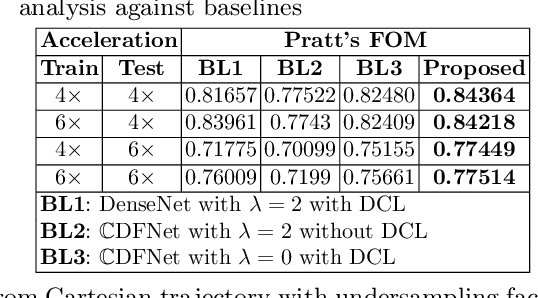
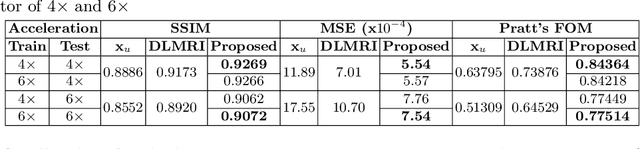
Abstract:Undersampling the k-space data is widely adopted for acceleration of Magnetic Resonance Imaging (MRI). Current deep learning based approaches for supervised learning of MRI image reconstruction employ real-valued operations and representations by treating complex valued k-space/spatial-space as real values. In this paper, we propose complex dense fully convolutional neural network ($\mathbb{C}$DFNet) for learning to de-alias the reconstruction artifacts within undersampled MRI images. We fashioned a densely-connected fully convolutional block tailored for complex-valued inputs by introducing dedicated layers such as complex convolution, batch normalization, non-linearities etc. $\mathbb{C}$DFNet leverages the inherently complex-valued nature of input k-space and learns richer representations. We demonstrate improved perceptual quality and recovery of anatomical structures through $\mathbb{C}$DFNet in contrast to its real-valued counterparts.
SynNet: Structure-Preserving Fully Convolutional Networks for Medical Image Synthesis
Jun 29, 2018



Abstract:Cross modal image syntheses is gaining significant interests for its ability to estimate target images of a different modality from a given set of source images,like estimating MR to MR, MR to CT, CT to PET etc, without the need for an actual acquisition.Though they show potential for applications in radiation therapy planning,image super resolution, atlas construction, image segmentation etc.The synthesis results are not as accurate as the actual acquisition.In this paper,we address the problem of multi modal image synthesis by proposing a fully convolutional deep learning architecture called the SynNet.We extend the proposed architecture for various input output configurations. And finally, we propose a structure preserving custom loss function for cross-modal image synthesis.We validate the proposed SynNet and its extended framework on BRATS dataset with comparisons against three state-of-the art methods.And the results of the proposed custom loss function is validated against the traditional loss function used by the state-of-the-art methods for cross modal image synthesis.
Inherent Brain Segmentation Quality Control from Fully ConvNet Monte Carlo Sampling
Jun 08, 2018



Abstract:We introduce inherent measures for effective quality control of brain segmentation based on a Bayesian fully convolutional neural network, using model uncertainty. Monte Carlo samples from the posterior distribution are efficiently generated using dropout at test time. Based on these samples, we introduce next to a voxel-wise uncertainty map also three metrics for structure-wise uncertainty. We then incorporate these structure-wise uncertainty in group analyses as a measure of confidence in the observation. Our results show that the metrics are highly correlated to segmentation accuracy and therefore present an inherent measure of segmentation quality. Furthermore, group analysis with uncertainty results in effect sizes closer to that of manual annotations. The introduced uncertainty metrics can not only be very useful in translation to clinical practice but also provide automated quality control and group analyses in processing large data repositories.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge