Tony Stöcker
Automated Olfactory Bulb Segmentation on High Resolutional T2-Weighted MRI
Aug 09, 2021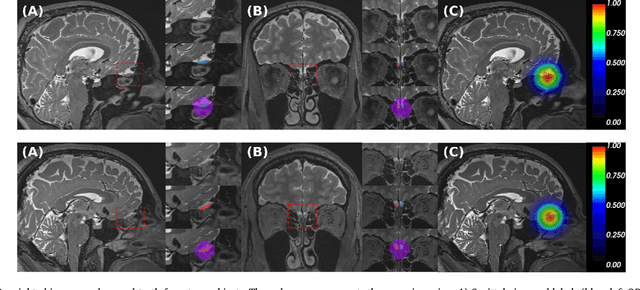
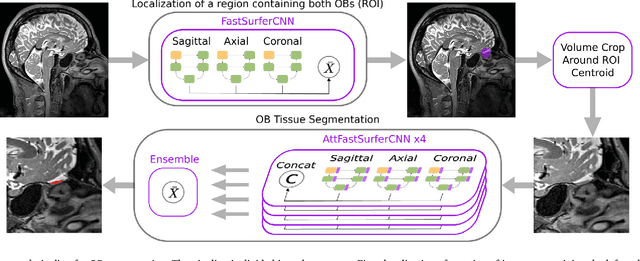
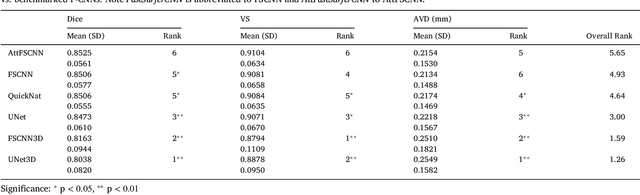
Abstract:The neuroimage analysis community has neglected the automated segmentation of the olfactory bulb (OB) despite its crucial role in olfactory function. The lack of an automatic processing method for the OB can be explained by its challenging properties. Nonetheless, recent advances in MRI acquisition techniques and resolution have allowed raters to generate more reliable manual annotations. Furthermore, the high accuracy of deep learning methods for solving semantic segmentation problems provides us with an option to reliably assess even small structures. In this work, we introduce a novel, fast, and fully automated deep learning pipeline to accurately segment OB tissue on sub-millimeter T2-weighted (T2w) whole-brain MR images. To this end, we designed a three-stage pipeline: (1) Localization of a region containing both OBs using FastSurferCNN, (2) Segmentation of OB tissue within the localized region through four independent AttFastSurferCNN - a novel deep learning architecture with a self-attention mechanism to improve modeling of contextual information, and (3) Ensemble of the predicted label maps. The OB pipeline exhibits high performance in terms of boundary delineation, OB localization, and volume estimation across a wide range of ages in 203 participants of the Rhineland Study. Moreover, it also generalizes to scans of an independent dataset never encountered during training, the Human Connectome Project (HCP), with different acquisition parameters and demographics, evaluated in 30 cases at the native 0.7mm HCP resolution, and the default 0.8mm pipeline resolution. We extensively validated our pipeline not only with respect to segmentation accuracy but also to known OB volume effects, where it can sensitively replicate age effects.
Complex Fully Convolutional Neural Networks for MR Image Reconstruction
Jul 09, 2018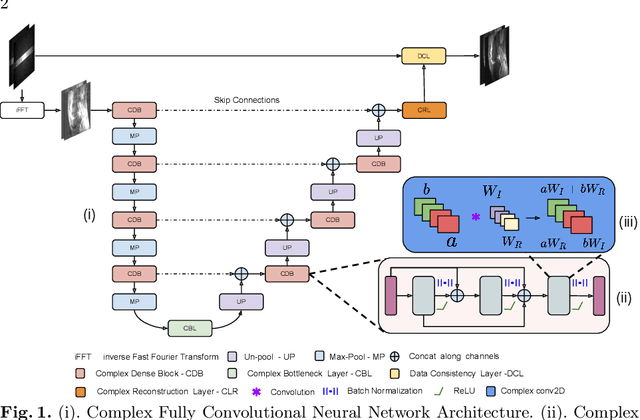
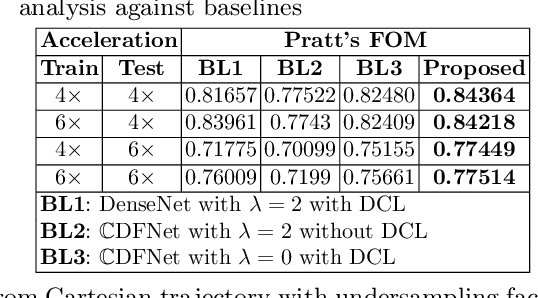
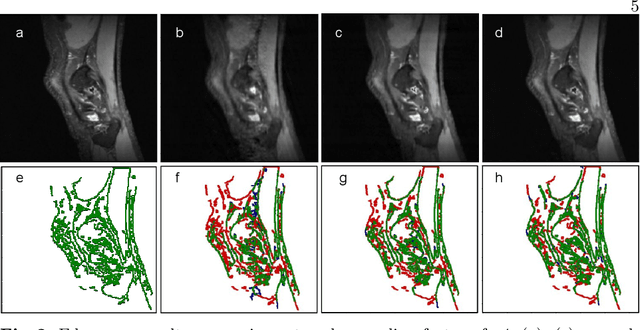
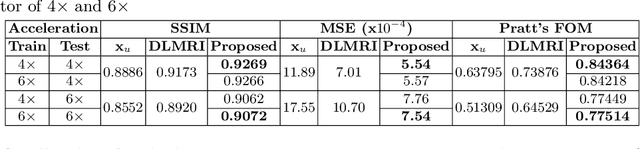
Abstract:Undersampling the k-space data is widely adopted for acceleration of Magnetic Resonance Imaging (MRI). Current deep learning based approaches for supervised learning of MRI image reconstruction employ real-valued operations and representations by treating complex valued k-space/spatial-space as real values. In this paper, we propose complex dense fully convolutional neural network ($\mathbb{C}$DFNet) for learning to de-alias the reconstruction artifacts within undersampled MRI images. We fashioned a densely-connected fully convolutional block tailored for complex-valued inputs by introducing dedicated layers such as complex convolution, batch normalization, non-linearities etc. $\mathbb{C}$DFNet leverages the inherently complex-valued nature of input k-space and learns richer representations. We demonstrate improved perceptual quality and recovery of anatomical structures through $\mathbb{C}$DFNet in contrast to its real-valued counterparts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge