Igor Yakushev
PASTA: Pathology-Aware MRI to PET Cross-Modal Translation with Diffusion Models
May 27, 2024Abstract:Positron emission tomography (PET) is a well-established functional imaging technique for diagnosing brain disorders. However, PET's high costs and radiation exposure limit its widespread use. In contrast, magnetic resonance imaging (MRI) does not have these limitations. Although it also captures neurodegenerative changes, MRI is a less sensitive diagnostic tool than PET. To close this gap, we aim to generate synthetic PET from MRI. Herewith, we introduce PASTA, a novel pathology-aware image translation framework based on conditional diffusion models. Compared to the state-of-the-art methods, PASTA excels in preserving both structural and pathological details in the target modality, which is achieved through its highly interactive dual-arm architecture and multi-modal condition integration. A cycle exchange consistency and volumetric generation strategy elevate PASTA's capability to produce high-quality 3D PET scans. Our qualitative and quantitative results confirm that the synthesized PET scans from PASTA not only reach the best quantitative scores but also preserve the pathology correctly. For Alzheimer's classification, the performance of synthesized scans improves over MRI by 4%, almost reaching the performance of actual PET. Code is available at https://github.com/ai-med/PASTA.
From Barlow Twins to Triplet Training: Differentiating Dementia with Limited Data
Apr 09, 2024Abstract:Differential diagnosis of dementia is challenging due to overlapping symptoms, with structural magnetic resonance imaging (MRI) being the primary method for diagnosis. Despite the clinical value of computer-aided differential diagnosis, research has been limited, mainly due to the absence of public datasets that contain diverse types of dementia. This leaves researchers with small in-house datasets that are insufficient for training deep neural networks (DNNs). Self-supervised learning shows promise for utilizing unlabeled MRI scans in training, but small batch sizes for volumetric brain scans make its application challenging. To address these issues, we propose Triplet Training for differential diagnosis with limited target data. It consists of three key stages: (i) self-supervised pre-training on unlabeled data with Barlow Twins, (ii) self-distillation on task-related data, and (iii) fine-tuning on the target dataset. Our approach significantly outperforms traditional training strategies, achieving a balanced accuracy of 75.6%. We further provide insights into the training process by visualizing changes in the latent space after each step. Finally, we validate the robustness of Triplet Training in terms of its individual components in a comprehensive ablation study. Our code is available at https://github.com/ai-med/TripletTraining.
Is a PET all you need? A multi-modal study for Alzheimer's disease using 3D CNNs
Jul 05, 2022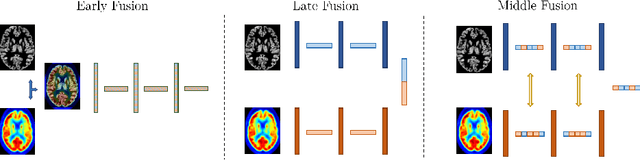
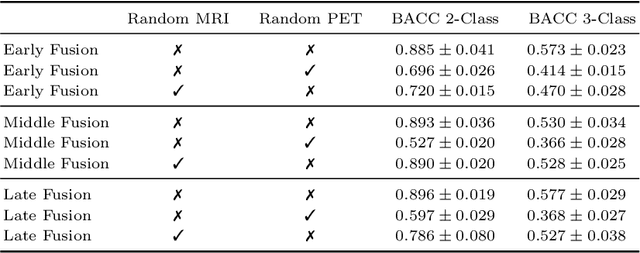
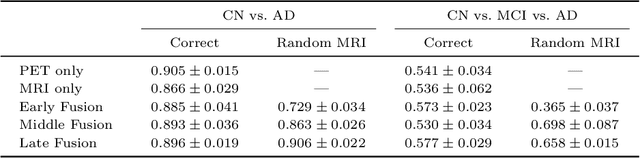
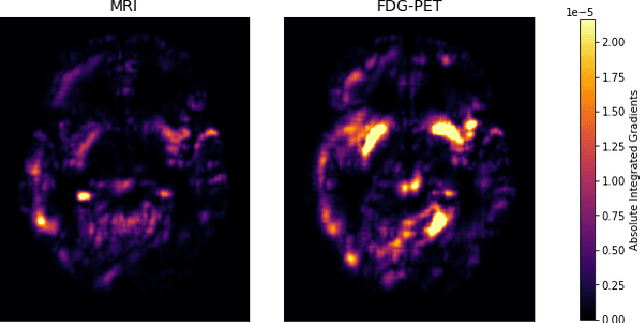
Abstract:Alzheimer's Disease (AD) is the most common form of dementia and often difficult to diagnose due to the multifactorial etiology of dementia. Recent works on neuroimaging-based computer-aided diagnosis with deep neural networks (DNNs) showed that fusing structural magnetic resonance images (sMRI) and fluorodeoxyglucose positron emission tomography (FDG-PET) leads to improved accuracy in a study population of healthy controls and subjects with AD. However, this result conflicts with the established clinical knowledge that FDG-PET better captures AD-specific pathologies than sMRI. Therefore, we propose a framework for the systematic evaluation of multi-modal DNNs and critically re-evaluate single- and multi-modal DNNs based on FDG-PET and sMRI for binary healthy vs. AD, and three-way healthy/mild cognitive impairment/AD classification. Our experiments demonstrate that a single-modality network using FDG-PET performs better than MRI (accuracy 0.91 vs 0.87) and does not show improvement when combined. This conforms with the established clinical knowledge on AD biomarkers, but raises questions about the true benefit of multi-modal DNNs. We argue that future work on multi-modal fusion should systematically assess the contribution of individual modalities following our proposed evaluation framework. Finally, we encourage the community to go beyond healthy vs. AD classification and focus on differential diagnosis of dementia, where fusing multi-modal image information conforms with a clinical need.
Learning Optimal Deep Projection of $^{18}$F-FDG PET Imaging for Early Differential Diagnosis of Parkinsonian Syndromes
Oct 11, 2018


Abstract:Several diseases of parkinsonian syndromes present similar symptoms at early stage and no objective widely used diagnostic methods have been approved until now. Positron emission tomography (PET) with $^{18}$F-FDG was shown to be able to assess early neuronal dysfunction of synucleinopathies and tauopathies. Tensor factorization (TF) based approaches have been applied to identify characteristic metabolic patterns for differential diagnosis. However, these conventional dimension-reduction strategies assume linear or multi-linear relationships inside data, and are therefore insufficient to distinguish nonlinear metabolic differences between various parkinsonian syndromes. In this paper, we propose a Deep Projection Neural Network (DPNN) to identify characteristic metabolic pattern for early differential diagnosis of parkinsonian syndromes. We draw our inspiration from the existing TF methods. The network consists of a (i) compression part: which uses a deep network to learn optimal 2D projections of 3D scans, and a (ii) classification part: which maps the 2D projections to labels. The compression part can be pre-trained using surplus unlabelled datasets. Also, as the classification part operates on these 2D projections, it can be trained end-to-end effectively with limited labelled data, in contrast to 3D approaches. We show that DPNN is more effective in comparison to existing state-of-the-art and plausible baselines.
* 8 pages, 3 figures, conference, MICCAI DLMIA, 2018
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge