Ruogu Fang
Ouroboros: Single-step Diffusion Models for Cycle-consistent Forward and Inverse Rendering
Aug 20, 2025



Abstract:While multi-step diffusion models have advanced both forward and inverse rendering, existing approaches often treat these problems independently, leading to cycle inconsistency and slow inference speed. In this work, we present Ouroboros, a framework composed of two single-step diffusion models that handle forward and inverse rendering with mutual reinforcement. Our approach extends intrinsic decomposition to both indoor and outdoor scenes and introduces a cycle consistency mechanism that ensures coherence between forward and inverse rendering outputs. Experimental results demonstrate state-of-the-art performance across diverse scenes while achieving substantially faster inference speed compared to other diffusion-based methods. We also demonstrate that Ouroboros can transfer to video decomposition in a training-free manner, reducing temporal inconsistency in video sequences while maintaining high-quality per-frame inverse rendering.
OTSurv: A Novel Multiple Instance Learning Framework for Survival Prediction with Heterogeneity-aware Optimal Transport
Jun 25, 2025Abstract:Survival prediction using whole slide images (WSIs) can be formulated as a multiple instance learning (MIL) problem. However, existing MIL methods often fail to explicitly capture pathological heterogeneity within WSIs, both globally -- through long-tailed morphological distributions, and locally through -- tile-level prediction uncertainty. Optimal transport (OT) provides a principled way of modeling such heterogeneity by incorporating marginal distribution constraints. Building on this insight, we propose OTSurv, a novel MIL framework from an optimal transport perspective. Specifically, OTSurv formulates survival predictions as a heterogeneity-aware OT problem with two constraints: (1) global long-tail constraint that models prior morphological distributions to avert both mode collapse and excessive uniformity by regulating transport mass allocation, and (2) local uncertainty-aware constraint that prioritizes high-confidence patches while suppressing noise by progressively raising the total transport mass. We then recast the initial OT problem, augmented by these constraints, into an unbalanced OT formulation that can be solved with an efficient, hardware-friendly matrix scaling algorithm. Empirically, OTSurv sets new state-of-the-art results across six popular benchmarks, achieving an absolute 3.6% improvement in average C-index. In addition, OTSurv achieves statistical significance in log-rank tests and offers high interpretability, making it a powerful tool for survival prediction in digital pathology. Our codes are available at https://github.com/Y-Research-SBU/OTSurv.
PET Image Denoising via Text-Guided Diffusion: Integrating Anatomical Priors through Text Prompts
Feb 28, 2025Abstract:Low-dose Positron Emission Tomography (PET) imaging presents a significant challenge due to increased noise and reduced image quality, which can compromise its diagnostic accuracy and clinical utility. Denoising diffusion probabilistic models (DDPMs) have demonstrated promising performance for PET image denoising. However, existing DDPM-based methods typically overlook valuable metadata such as patient demographics, anatomical information, and scanning parameters, which should further enhance the denoising performance if considered. Recent advances in vision-language models (VLMs), particularly the pre-trained Contrastive Language-Image Pre-training (CLIP) model, have highlighted the potential of incorporating text-based information into visual tasks to improve downstream performance. In this preliminary study, we proposed a novel text-guided DDPM for PET image denoising that integrated anatomical priors through text prompts. Anatomical text descriptions were encoded using a pre-trained CLIP text encoder to extract semantic guidance, which was then incorporated into the diffusion process via the cross-attention mechanism. Evaluations based on paired 1/20 low-dose and normal-dose 18F-FDG PET datasets demonstrated that the proposed method achieved better quantitative performance than conventional UNet and standard DDPM methods at both the whole-body and organ levels. These results underscored the potential of leveraging VLMs to integrate rich metadata into the diffusion framework to enhance the image quality of low-dose PET scans.
Paleoinspired Vision: From Exploring Colour Vision Evolution to Inspiring Camera Design
Dec 27, 2024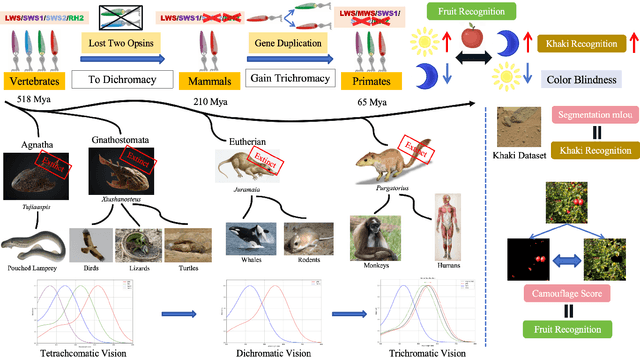
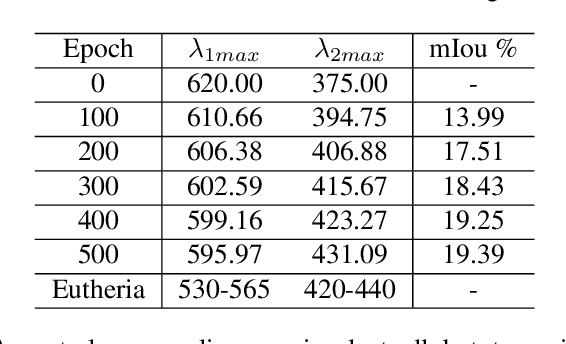
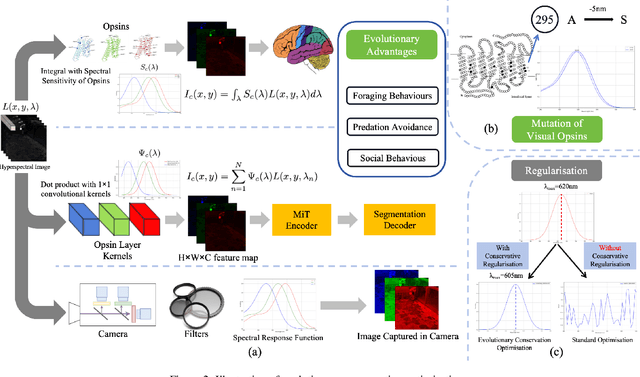
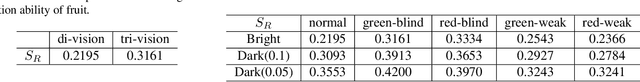
Abstract:The evolution of colour vision is captivating, as it reveals the adaptive strategies of extinct species while simultaneously inspiring innovations in modern imaging technology. In this study, we present a simplified model of visual transduction in the retina, introducing a novel opsin layer. We quantify evolutionary pressures by measuring machine vision recognition accuracy on colour images shaped by specific opsins. Building on this, we develop an evolutionary conservation optimisation algorithm to reconstruct the spectral sensitivity of opsins, enabling mutation-driven adaptations to to more effectively spot fruits or predators. This model condenses millions of years of evolution within seconds on GPU, providing an experimental framework to test long-standing hypotheses in evolutionary biology , such as vision of early mammals, primate trichromacy from gene duplication, retention of colour blindness, blue-shift of fish rod and multiple rod opsins with bioluminescence. Moreover, the model enables speculative explorations of hypothetical species, such as organisms with eyes adapted to the conditions on Mars. Our findings suggest a minimalist yet effective approach to task-specific camera filter design, optimising the spectral response function to meet application-driven demands. The code will be made publicly available upon acceptance.
A Comprehensive Survey of Foundation Models in Medicine
Jun 15, 2024Abstract:Foundation models (FMs) are large-scale deep-learning models trained on extensive datasets using self-supervised techniques. These models serve as a base for various downstream tasks, including healthcare. FMs have been adopted with great success across various domains within healthcare, including natural language processing (NLP), computer vision, graph learning, biology, and omics. Existing healthcare-based surveys have not yet included all of these domains. Therefore, this survey provides a comprehensive overview of FMs in healthcare. We focus on the history, learning strategies, flagship models, applications, and challenges of FMs. We explore how FMs such as the BERT and GPT families are reshaping various healthcare domains, including clinical large language models, medical image analysis, and omics data. Furthermore, we provide a detailed taxonomy of healthcare applications facilitated by FMs, such as clinical NLP, medical computer vision, graph learning, and other biology-related tasks. Despite the promising opportunities FMs provide, they also have several associated challenges, which are explained in detail. We also outline potential future directions to provide researchers and practitioners with insights into the potential and limitations of FMs in healthcare to advance their deployment and mitigate associated risks.
BrainFounder: Towards Brain Foundation Models for Neuroimage Analysis
Jun 14, 2024



Abstract:The burgeoning field of brain health research increasingly leverages artificial intelligence (AI) to interpret and analyze neurological data. This study introduces a novel approach towards the creation of medical foundation models by integrating a large-scale multi-modal magnetic resonance imaging (MRI) dataset derived from 41,400 participants in its own. Our method involves a novel two-stage pretraining approach using vision transformers. The first stage is dedicated to encoding anatomical structures in generally healthy brains, identifying key features such as shapes and sizes of different brain regions. The second stage concentrates on spatial information, encompassing aspects like location and the relative positioning of brain structures. We rigorously evaluate our model, BrainFounder, using the Brain Tumor Segmentation (BraTS) challenge and Anatomical Tracings of Lesions After Stroke v2.0 (ATLAS v2.0) datasets. BrainFounder demonstrates a significant performance gain, surpassing the achievements of the previous winning solutions using fully supervised learning. Our findings underscore the impact of scaling up both the complexity of the model and the volume of unlabeled training data derived from generally healthy brains, which enhances the accuracy and predictive capabilities of the model in complex neuroimaging tasks with MRI. The implications of this research provide transformative insights and practical applications in healthcare and make substantial steps towards the creation of foundation models for Medical AI. Our pretrained models and training code can be found at https://github.com/lab-smile/GatorBrain.
Morphological Profiling for Drug Discovery in the Era of Deep Learning
Dec 13, 2023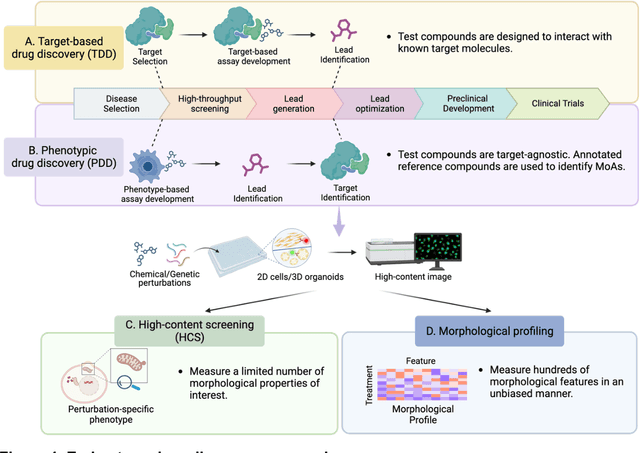
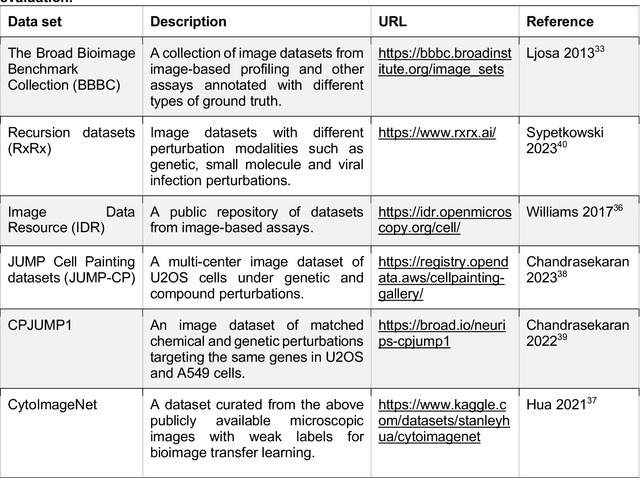
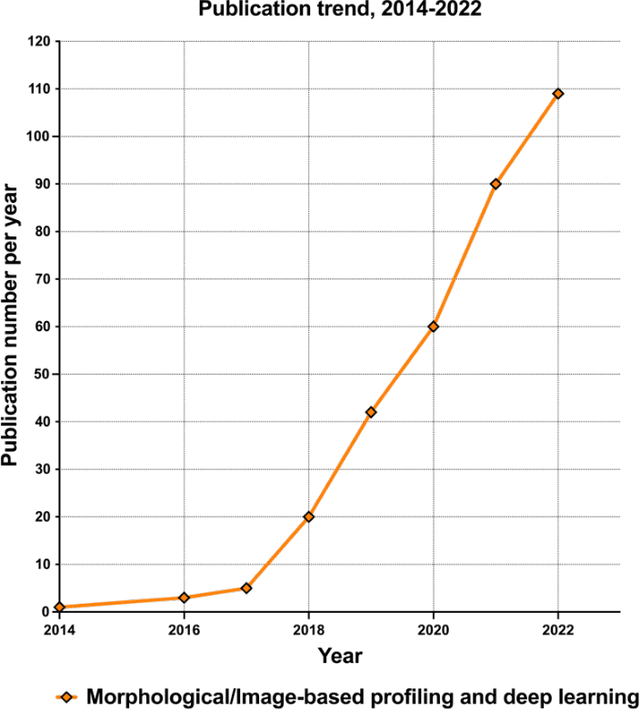
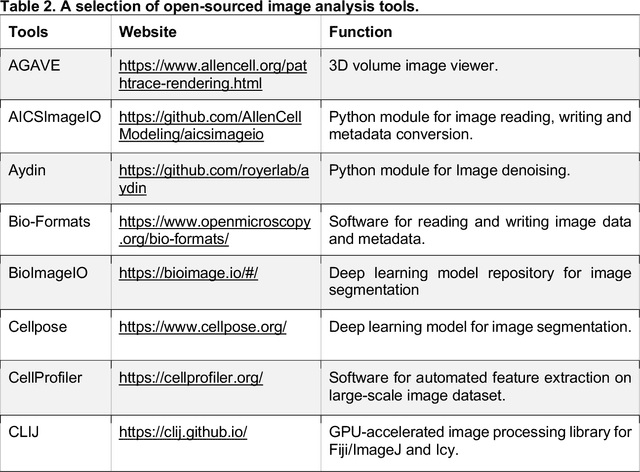
Abstract:Morphological profiling is a valuable tool in phenotypic drug discovery. The advent of high-throughput automated imaging has enabled the capturing of a wide range of morphological features of cells or organisms in response to perturbations at the single-cell resolution. Concurrently, significant advances in machine learning and deep learning, especially in computer vision, have led to substantial improvements in analyzing large-scale high-content images at high-throughput. These efforts have facilitated understanding of compound mechanism-of-action (MOA), drug repurposing, characterization of cell morphodynamics under perturbation, and ultimately contributing to the development of novel therapeutics. In this review, we provide a comprehensive overview of the recent advances in the field of morphological profiling. We summarize the image profiling analysis workflow, survey a broad spectrum of analysis strategies encompassing feature engineering- and deep learning-based approaches, and introduce publicly available benchmark datasets. We place a particular emphasis on the application of deep learning in this pipeline, covering cell segmentation, image representation learning, and multimodal learning. Additionally, we illuminate the application of morphological profiling in phenotypic drug discovery and highlight potential challenges and opportunities in this field.
DOMINO++: Domain-aware Loss Regularization for Deep Learning Generalizability
Aug 21, 2023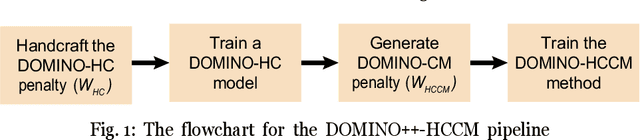
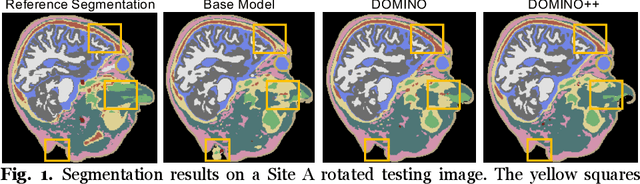
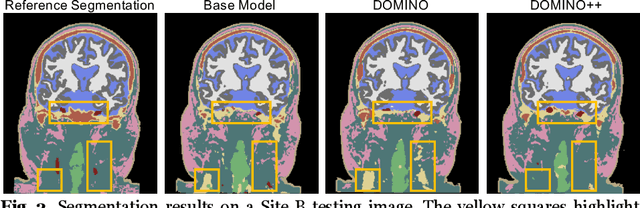

Abstract:Out-of-distribution (OOD) generalization poses a serious challenge for modern deep learning (DL). OOD data consists of test data that is significantly different from the model's training data. DL models that perform well on in-domain test data could struggle on OOD data. Overcoming this discrepancy is essential to the reliable deployment of DL. Proper model calibration decreases the number of spurious connections that are made between model features and class outputs. Hence, calibrated DL can improve OOD generalization by only learning features that are truly indicative of the respective classes. Previous work proposed domain-aware model calibration (DOMINO) to improve DL calibration, but it lacks designs for model generalizability to OOD data. In this work, we propose DOMINO++, a dual-guidance and dynamic domain-aware loss regularization focused on OOD generalizability. DOMINO++ integrates expert-guided and data-guided knowledge in its regularization. Unlike DOMINO which imposed a fixed scaling and regularization rate, DOMINO++ designs a dynamic scaling factor and an adaptive regularization rate. Comprehensive evaluations compare DOMINO++ with DOMINO and the baseline model for head tissue segmentation from magnetic resonance images (MRIs) on OOD data. The OOD data consists of synthetic noisy and rotated datasets, as well as real data using a different MRI scanner from a separate site. DOMINO++'s superior performance demonstrates its potential to improve the trustworthy deployment of DL on real clinical data.
Deep Learning Predicts Prevalent and Incident Parkinson's Disease From UK Biobank Fundus Imaging
Feb 17, 2023Abstract:Parkinson's disease is the world's fastest growing neurological disorder. Research to elucidate the mechanisms of Parkinson's disease and automate diagnostics would greatly improve the treatment of patients with Parkinson's disease. Current diagnostic methods are expensive with limited availability. Considering the long progression time of Parkinson's disease, a desirable screening should be diagnostically accurate even before the onset of symptoms to allow medical intervention. We promote attention for retinal fundus imaging, often termed a window to the brain, as a diagnostic screening modality for Parkinson's disease. We conduct a systematic evaluation of conventional machine learning and deep learning techniques to classify Parkinson's disease from UK Biobank fundus imaging. Our results suggest Parkinson's disease individuals can be differentiated from age and gender matched healthy subjects with 71% accuracy. This accuracy is maintained when predicting either prevalent or incident Parkinson's disease. Explainability and trustworthiness is enhanced by visual attribution maps of localized biomarkers and quantified metrics of model robustness to data perturbations.
DOMINO: Domain-aware Loss for Deep Learning Calibration
Feb 10, 2023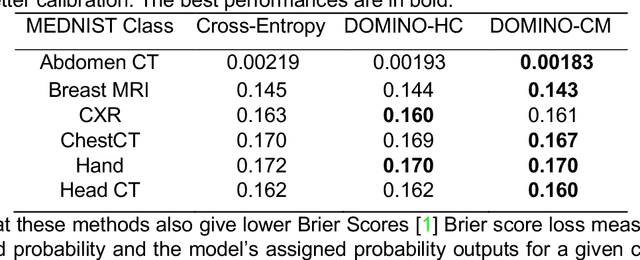
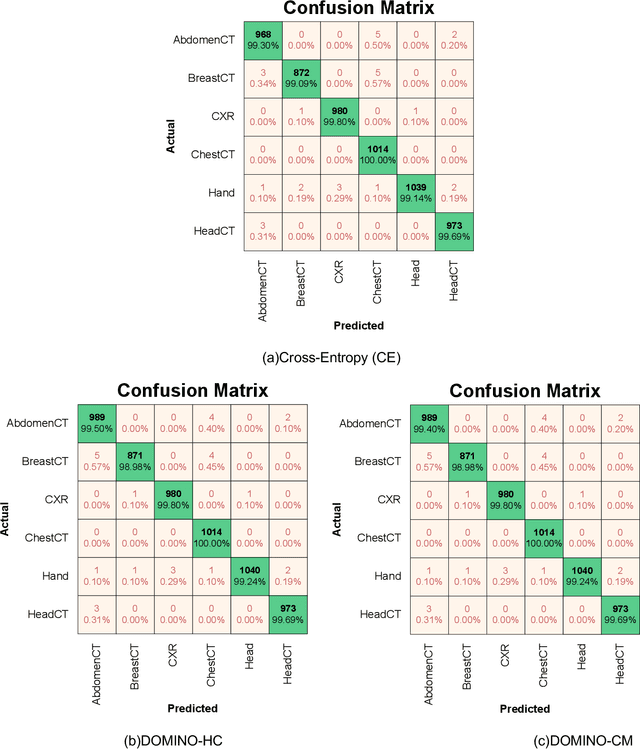
Abstract:Deep learning has achieved the state-of-the-art performance across medical imaging tasks; however, model calibration is often not considered. Uncalibrated models are potentially dangerous in high-risk applications since the user does not know when they will fail. Therefore, this paper proposes a novel domain-aware loss function to calibrate deep learning models. The proposed loss function applies a class-wise penalty based on the similarity between classes within a given target domain. Thus, the approach improves the calibration while also ensuring that the model makes less risky errors even when incorrect. The code for this software is available at https://github.com/lab-smile/DOMINO.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge