Francesca De Benetti
AutoPaint: A Self-Inpainting Method for Unsupervised Anomaly Detection
May 21, 2023Abstract:Robust and accurate detection and segmentation of heterogenous tumors appearing in different anatomical organs with supervised methods require large-scale labeled datasets covering all possible types of diseases. Due to the unavailability of such rich datasets and the high cost of annotations, unsupervised anomaly detection (UAD) methods have been developed aiming to detect the pathologies as deviation from the normality by utilizing the unlabeled healthy image data. However, developed UAD models are often trained with an incomplete distribution of healthy anatomies and have difficulties in preserving anatomical constraints. This work intends to, first, propose a robust inpainting model to learn the details of healthy anatomies and reconstruct high-resolution images by preserving anatomical constraints. Second, we propose an autoinpainting pipeline to automatically detect tumors, replace their appearance with the learned healthy anatomies, and based on that segment the tumoral volumes in a purely unsupervised fashion. Three imaging datasets, including PET, CT, and PET-CT scans of lung tumors and head and neck tumors, are studied as benchmarks for evaluation. Experimental results demonstrate the significant superiority of the proposed method over a wide range of state-of-the-art UAD methods. Moreover, the unsupervised method we propose produces comparable results to a robust supervised segmentation method when applied to multimodal images.
Self-Supervised Learning for Physiologically-Based Pharmacokinetic Modeling in Dynamic PET
May 17, 2023



Abstract:Dynamic positron emission tomography imaging (dPET) provides temporally resolved images of a tracer enabling a quantitative measure of physiological processes. Voxel-wise physiologically-based pharmacokinetic (PBPK) modeling of the time activity curves (TAC) can provide relevant diagnostic information for clinical workflow. Conventional fitting strategies for TACs are slow and ignore the spatial relation between neighboring voxels. We train a spatio-temporal UNet to estimate the kinetic parameters given TAC from F-18-fluorodeoxyglucose (FDG) dPET. This work introduces a self-supervised loss formulation to enforce the similarity between the measured TAC and those generated with the learned kinetic parameters. Our method provides quantitatively comparable results at organ-level to the significantly slower conventional approaches, while generating pixel-wise parametric images which are consistent with expected physiology. To the best of our knowledge, this is the first self-supervised network that allows voxel-wise computation of kinetic parameters consistent with a non-linear kinetic model. The code will become publicly available upon acceptance.
Weakly-supervised Biomechanically-constrained CT/MRI Registration of the Spine
May 16, 2022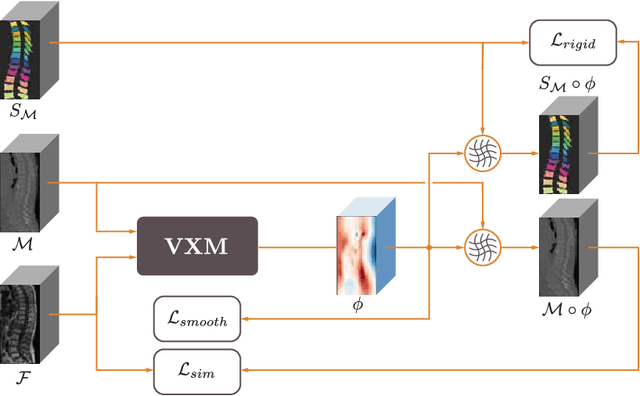
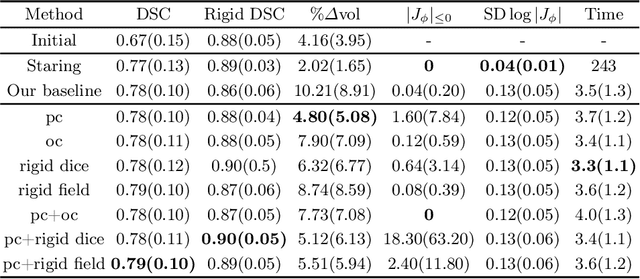
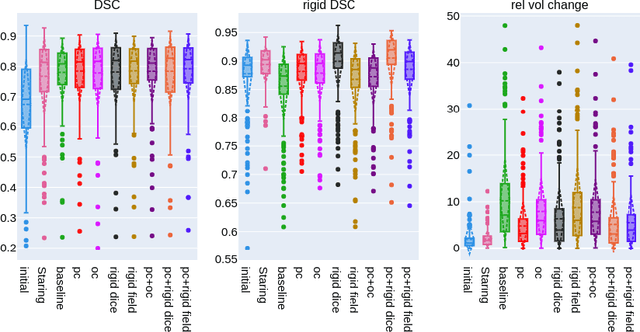
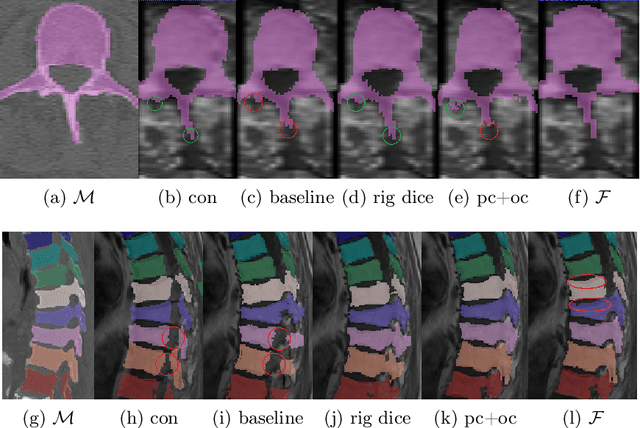
Abstract:CT and MRI are two of the most informative modalities in spinal diagnostics and treatment planning. CT is useful when analysing bony structures, while MRI gives information about the soft tissue. Thus, fusing the information of both modalities can be very beneficial. Registration is the first step for this fusion. While the soft tissues around the vertebra are deformable, each vertebral body is constrained to move rigidly. We propose a weakly-supervised deep learning framework that preserves the rigidity and the volume of each vertebra while maximizing the accuracy of the registration. To achieve this goal, we introduce anatomy-aware losses for training the network. We specifically design these losses to depend only on the CT label maps since automatic vertebra segmentation in CT gives more accurate results contrary to MRI. We evaluate our method on an in-house dataset of 167 patients. Our results show that adding the anatomy-aware losses increases the plausibility of the inferred transformation while keeping the accuracy untouched.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021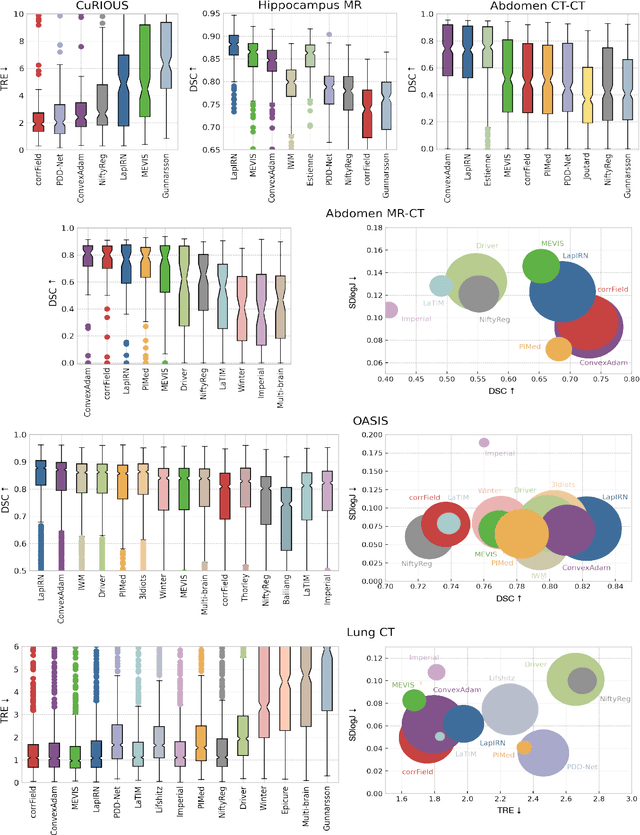
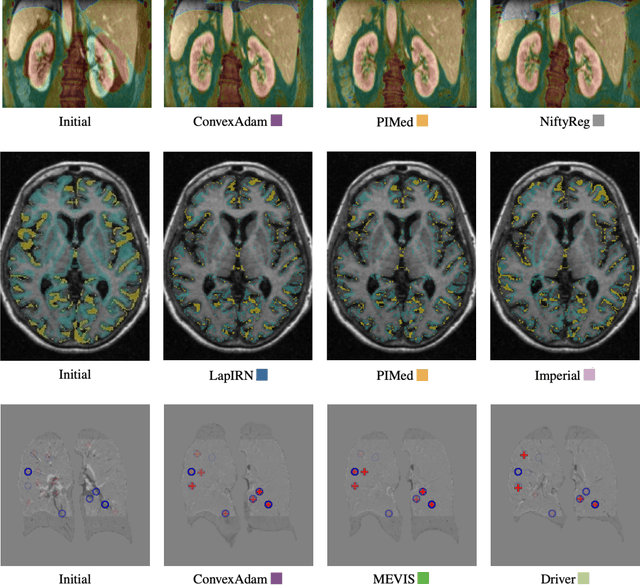
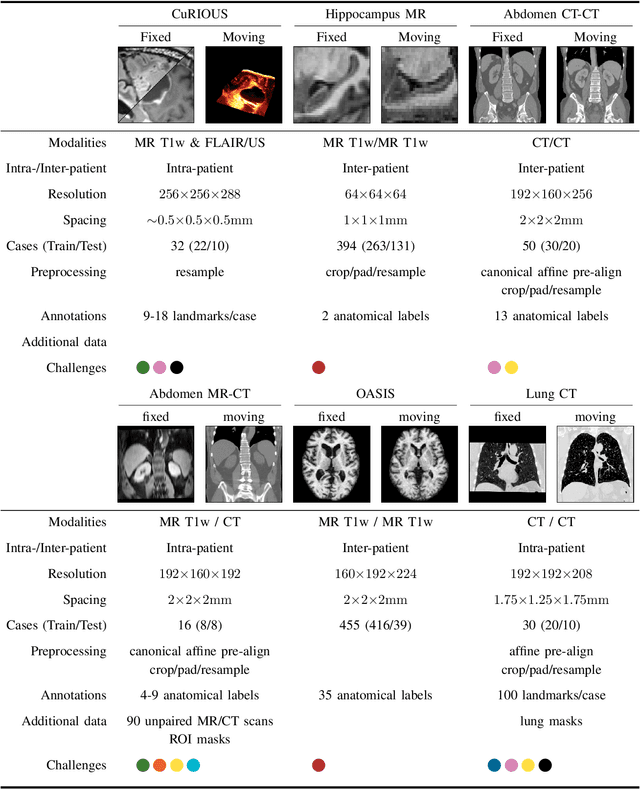
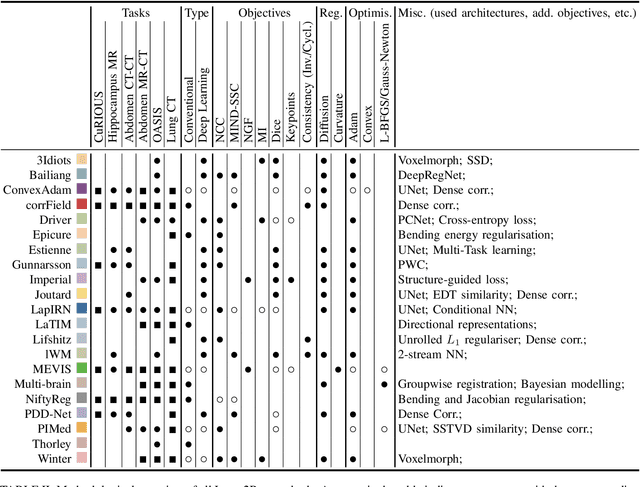
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
Patient-specific virtual spine straightening and vertebra inpainting: An automatic framework for osteoplasty planning
Mar 23, 2021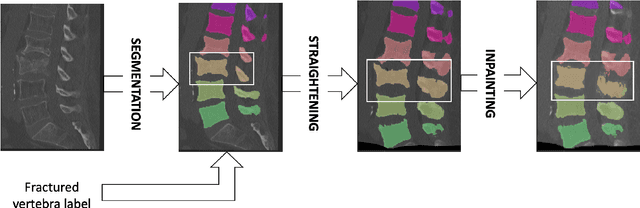

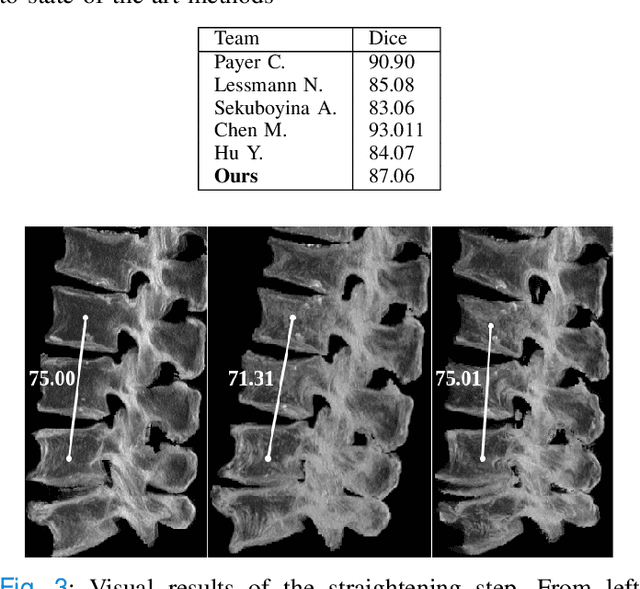
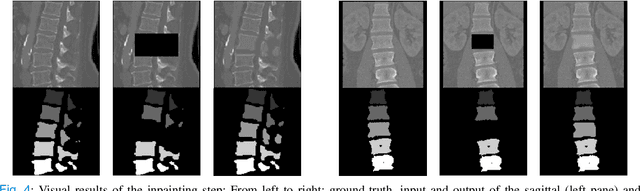
Abstract:Symptomatic spinal vertebral compression fractures (VCFs) often require osteoplasty treatment. A cement-like material is injected into the bone to stabilize the fracture, restore the vertebral body height and alleviate pain. Leakage is a common complication and may occur due to too much cement being injected. In this work, we propose an automated patient-specific framework that can allow physicians to calculate an upper bound of cement for the injection and estimate the optimal outcome of osteoplasty. The framework uses the patient CT scan and the fractured vertebra label to build a virtual healthy spine using a high-level approach. Firstly, the fractured spine is segmented with a three-step Convolution Neural Network (CNN) architecture. Next, a per-vertebra rigid registration to a healthy spine atlas restores its curvature. Finally, a GAN-based inpainting approach replaces the fractured vertebra with an estimation of its original shape. Based on this outcome, we then estimate the maximum amount of bone cement for injection. We evaluate our framework by comparing the virtual vertebrae volumes of ten patients to their healthy equivalent and report an average error of 3.88$\pm$7.63\%. The presented pipeline offers a first approach to a personalized automatic high-level framework for planning osteoplasty procedures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge