Lasse Hansen
Beyond the LUMIR challenge: The pathway to foundational registration models
May 30, 2025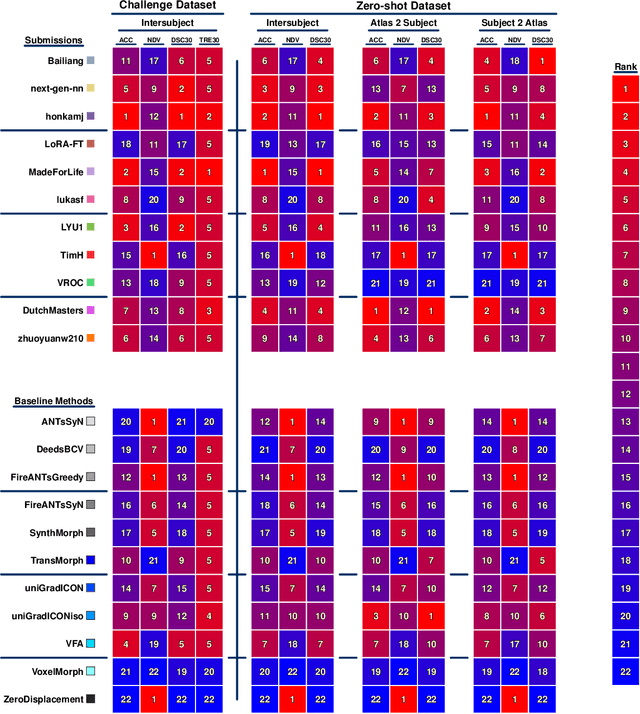
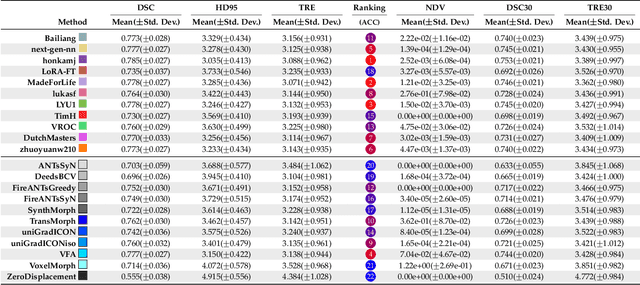
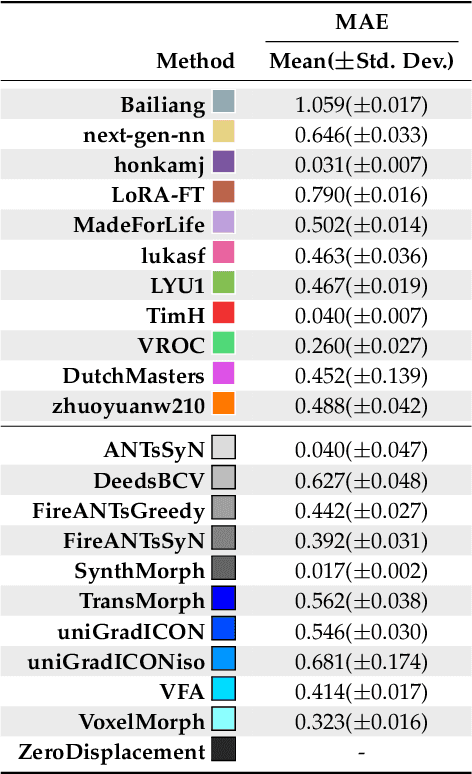
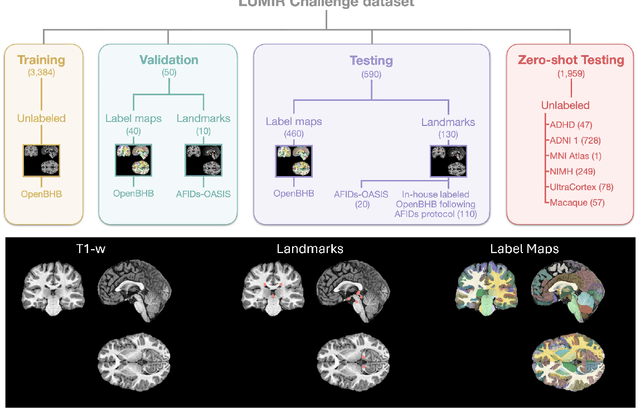
Abstract:Medical image challenges have played a transformative role in advancing the field, catalyzing algorithmic innovation and establishing new performance standards across diverse clinical applications. Image registration, a foundational task in neuroimaging pipelines, has similarly benefited from the Learn2Reg initiative. Building on this foundation, we introduce the Large-scale Unsupervised Brain MRI Image Registration (LUMIR) challenge, a next-generation benchmark designed to assess and advance unsupervised brain MRI registration. Distinct from prior challenges that leveraged anatomical label maps for supervision, LUMIR removes this dependency by providing over 4,000 preprocessed T1-weighted brain MRIs for training without any label maps, encouraging biologically plausible deformation modeling through self-supervision. In addition to evaluating performance on 590 held-out test subjects, LUMIR introduces a rigorous suite of zero-shot generalization tasks, spanning out-of-domain imaging modalities (e.g., FLAIR, T2-weighted, T2*-weighted), disease populations (e.g., Alzheimer's disease), acquisition protocols (e.g., 9.4T MRI), and species (e.g., macaque brains). A total of 1,158 subjects and over 4,000 image pairs were included for evaluation. Performance was assessed using both segmentation-based metrics (Dice coefficient, 95th percentile Hausdorff distance) and landmark-based registration accuracy (target registration error). Across both in-domain and zero-shot tasks, deep learning-based methods consistently achieved state-of-the-art accuracy while producing anatomically plausible deformation fields. The top-performing deep learning-based models demonstrated diffeomorphic properties and inverse consistency, outperforming several leading optimization-based methods, and showing strong robustness to most domain shifts, the exception being a drop in performance on out-of-domain contrasts.
OncoReg: Medical Image Registration for Oncological Challenges
Apr 01, 2025Abstract:In modern cancer research, the vast volume of medical data generated is often underutilised due to challenges related to patient privacy. The OncoReg Challenge addresses this issue by enabling researchers to develop and validate image registration methods through a two-phase framework that ensures patient privacy while fostering the development of more generalisable AI models. Phase one involves working with a publicly available dataset, while phase two focuses on training models on a private dataset within secure hospital networks. OncoReg builds upon the foundation established by the Learn2Reg Challenge by incorporating the registration of interventional cone-beam computed tomography (CBCT) with standard planning fan-beam CT (FBCT) images in radiotherapy. Accurate image registration is crucial in oncology, particularly for dynamic treatment adjustments in image-guided radiotherapy, where precise alignment is necessary to minimise radiation exposure to healthy tissues while effectively targeting tumours. This work details the methodology and data behind the OncoReg Challenge and provides a comprehensive analysis of the competition entries and results. Findings reveal that feature extraction plays a pivotal role in this registration task. A new method emerging from this challenge demonstrated its versatility, while established approaches continue to perform comparably to newer techniques. Both deep learning and classical approaches still play significant roles in image registration, with the combination of methods - particularly in feature extraction - proving most effective.
MMTEB: Massive Multilingual Text Embedding Benchmark
Feb 19, 2025Abstract:Text embeddings are typically evaluated on a limited set of tasks, which are constrained by language, domain, and task diversity. To address these limitations and provide a more comprehensive evaluation, we introduce the Massive Multilingual Text Embedding Benchmark (MMTEB) - a large-scale, community-driven expansion of MTEB, covering over 500 quality-controlled evaluation tasks across 250+ languages. MMTEB includes a diverse set of challenging, novel tasks such as instruction following, long-document retrieval, and code retrieval, representing the largest multilingual collection of evaluation tasks for embedding models to date. Using this collection, we develop several highly multilingual benchmarks, which we use to evaluate a representative set of models. We find that while large language models (LLMs) with billions of parameters can achieve state-of-the-art performance on certain language subsets and task categories, the best-performing publicly available model is multilingual-e5-large-instruct with only 560 million parameters. To facilitate accessibility and reduce computational cost, we introduce a novel downsampling method based on inter-task correlation, ensuring a diverse selection while preserving relative model rankings. Furthermore, we optimize tasks such as retrieval by sampling hard negatives, creating smaller but effective splits. These optimizations allow us to introduce benchmarks that drastically reduce computational demands. For instance, our newly introduced zero-shot English benchmark maintains a ranking order similar to the full-scale version but at a fraction of the computational cost.
SAM Carries the Burden: A Semi-Supervised Approach Refining Pseudo Labels for Medical Segmentation
Nov 19, 2024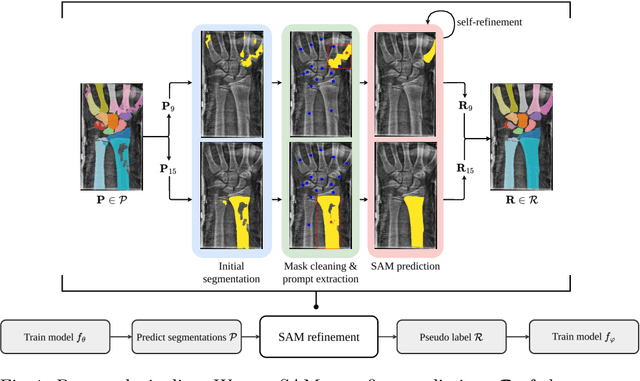
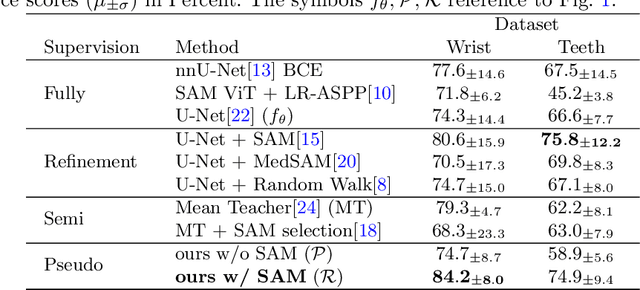
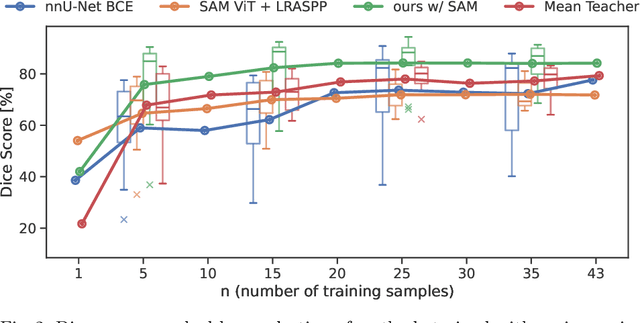
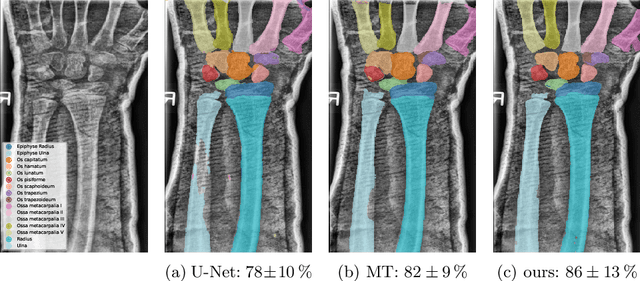
Abstract:Semantic segmentation is a crucial task in medical imaging. Although supervised learning techniques have proven to be effective in performing this task, they heavily depend on large amounts of annotated training data. The recently introduced Segment Anything Model (SAM) enables prompt-based segmentation and offers zero-shot generalization to unfamiliar objects. In our work, we leverage SAM's abstract object understanding for medical image segmentation to provide pseudo labels for semi-supervised learning, thereby mitigating the need for extensive annotated training data. Our approach refines initial segmentations that are derived from a limited amount of annotated data (comprising up to 43 cases) by extracting bounding boxes and seed points as prompts forwarded to SAM. Thus, it enables the generation of dense segmentation masks as pseudo labels for unlabelled data. The results show that training with our pseudo labels yields an improvement in Dice score from $74.29\,\%$ to $84.17\,\%$ and from $66.63\,\%$ to $74.87\,\%$ for the segmentation of bones of the paediatric wrist and teeth in dental radiographs, respectively. As a result, our method outperforms intensity-based post-processing methods, state-of-the-art supervised learning for segmentation (nnU-Net), and the semi-supervised mean teacher approach. Our Code is available on GitHub.
DenseSeg: Joint Learning for Semantic Segmentation and Landmark Detection Using Dense Image-to-Shape Representation
May 30, 2024



Abstract:Purpose: Semantic segmentation and landmark detection are fundamental tasks of medical image processing, facilitating further analysis of anatomical objects. Although deep learning-based pixel-wise classification has set a new-state-of-the-art for segmentation, it falls short in landmark detection, a strength of shape-based approaches. Methods: In this work, we propose a dense image-to-shape representation that enables the joint learning of landmarks and semantic segmentation by employing a fully convolutional architecture. Our method intuitively allows the extraction of arbitrary landmarks due to its representation of anatomical correspondences. We benchmark our method against the state-of-the-art for semantic segmentation (nnUNet), a shape-based approach employing geometric deep learning and a CNN-based method for landmark detection. Results: We evaluate our method on two medical dataset: one common benchmark featuring the lungs, heart, and clavicle from thorax X-rays, and another with 17 different bones in the paediatric wrist. While our method is on pair with the landmark detection baseline in the thorax setting (error in mm of $2.6\pm0.9$ vs $2.7\pm0.9$), it substantially surpassed it in the more complex wrist setting ($1.1\pm0.6$ vs $1.9\pm0.5$). Conclusion: We demonstrate that dense geometric shape representation is beneficial for challenging landmark detection tasks and outperforms previous state-of-the-art using heatmap regression. While it does not require explicit training on the landmarks themselves, allowing for the addition of new landmarks without necessitating retraining.}
Danish Foundation Models
Nov 13, 2023

Abstract:Large language models, sometimes referred to as foundation models, have transformed multiple fields of research. However, smaller languages risk falling behind due to high training costs and small incentives for large companies to train these models. To combat this, the Danish Foundation Models project seeks to provide and maintain open, well-documented, and high-quality foundation models for the Danish language. This is achieved through broad cooperation with public and private institutions, to ensure high data quality and applicability of the trained models. We present the motivation of the project, the current status, and future perspectives.
Reimagining Synthetic Tabular Data Generation through Data-Centric AI: A Comprehensive Benchmark
Oct 25, 2023



Abstract:Synthetic data serves as an alternative in training machine learning models, particularly when real-world data is limited or inaccessible. However, ensuring that synthetic data mirrors the complex nuances of real-world data is a challenging task. This paper addresses this issue by exploring the potential of integrating data-centric AI techniques which profile the data to guide the synthetic data generation process. Moreover, we shed light on the often ignored consequences of neglecting these data profiles during synthetic data generation -- despite seemingly high statistical fidelity. Subsequently, we propose a novel framework to evaluate the integration of data profiles to guide the creation of more representative synthetic data. In an empirical study, we evaluate the performance of five state-of-the-art models for tabular data generation on eleven distinct tabular datasets. The findings offer critical insights into the successes and limitations of current synthetic data generation techniques. Finally, we provide practical recommendations for integrating data-centric insights into the synthetic data generation process, with a specific focus on classification performance, model selection, and feature selection. This study aims to reevaluate conventional approaches to synthetic data generation and promote the application of data-centric AI techniques in improving the quality and effectiveness of synthetic data.
Unsupervised 3D registration through optimization-guided cyclical self-training
Jun 29, 2023



Abstract:State-of-the-art deep learning-based registration methods employ three different learning strategies: supervised learning, which requires costly manual annotations, unsupervised learning, which heavily relies on hand-crafted similarity metrics designed by domain experts, or learning from synthetic data, which introduces a domain shift. To overcome the limitations of these strategies, we propose a novel self-supervised learning paradigm for unsupervised registration, relying on self-training. Our idea is based on two key insights. Feature-based differentiable optimizers 1) perform reasonable registration even from random features and 2) stabilize the training of the preceding feature extraction network on noisy labels. Consequently, we propose cyclical self-training, where pseudo labels are initialized as the displacement fields inferred from random features and cyclically updated based on more and more expressive features from the learning feature extractor, yielding a self-reinforcement effect. We evaluate the method for abdomen and lung registration, consistently surpassing metric-based supervision and outperforming diverse state-of-the-art competitors. Source code is available at https://github.com/multimodallearning/reg-cyclical-self-train.
Why is the winner the best?
Mar 30, 2023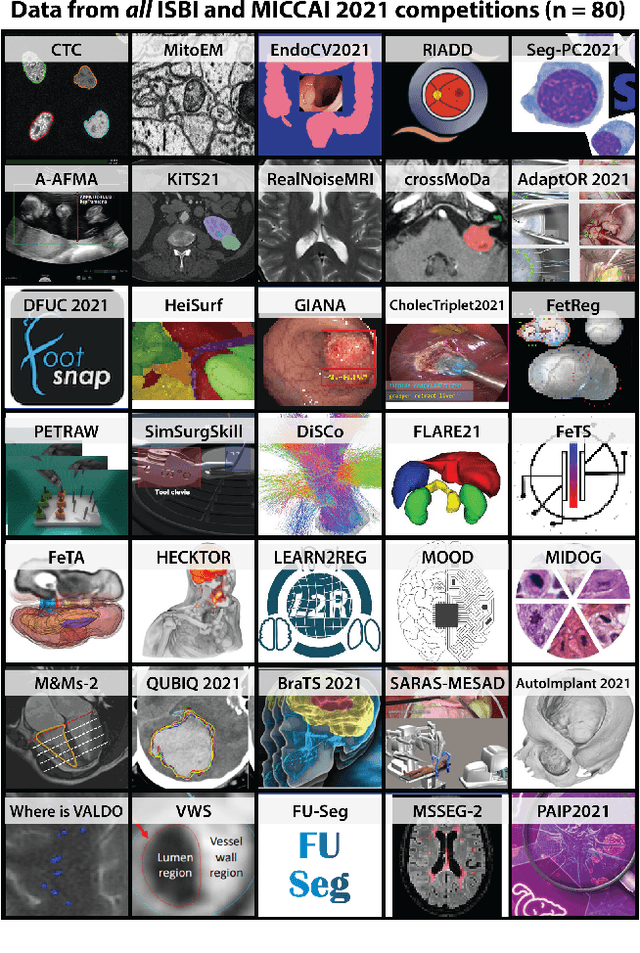
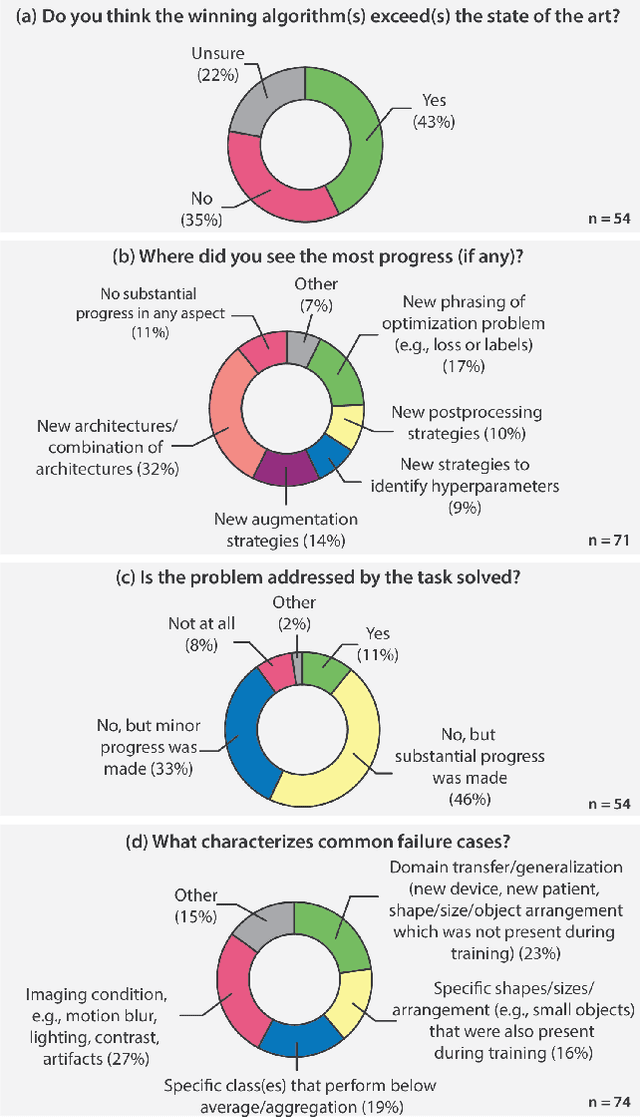
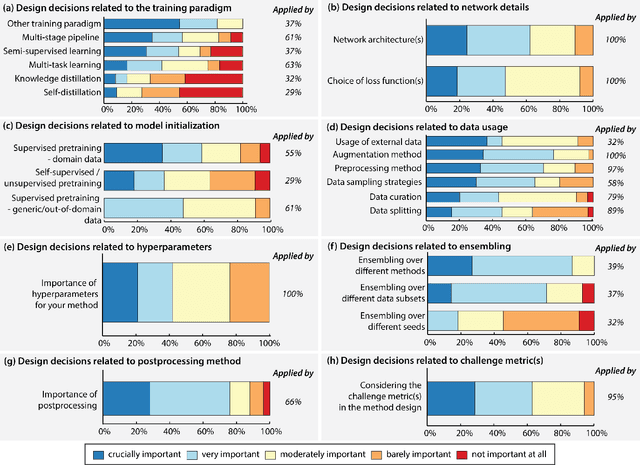
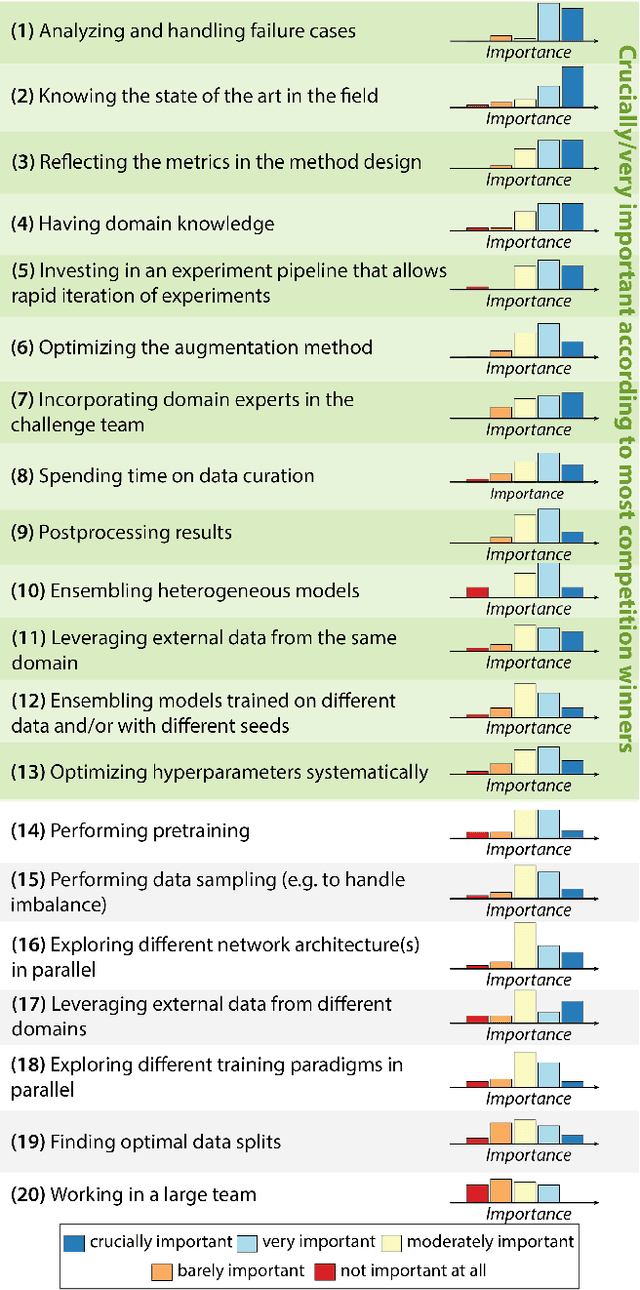
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Automated speech- and text-based classification of neuropsychiatric conditions in a multidiagnostic setting
Jan 13, 2023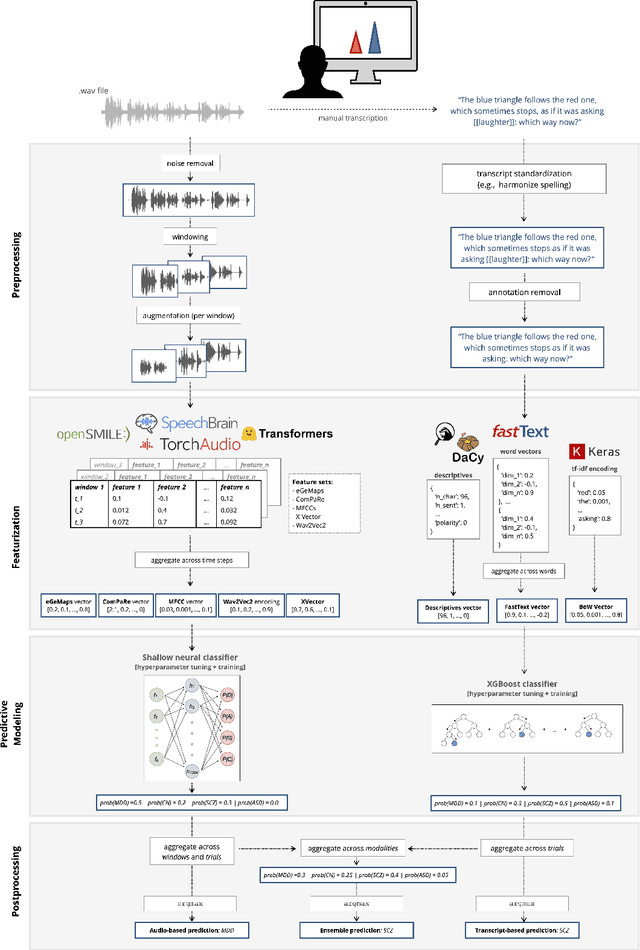
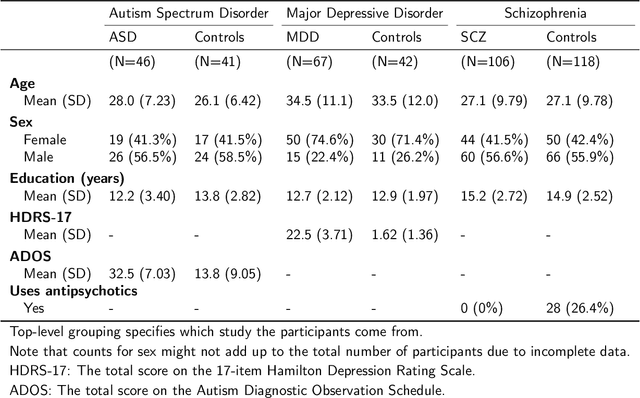
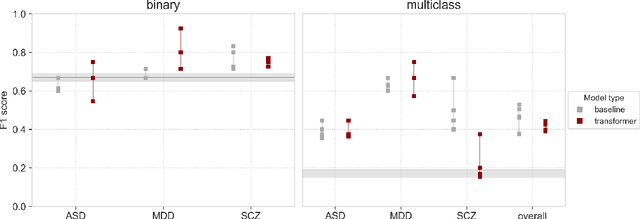
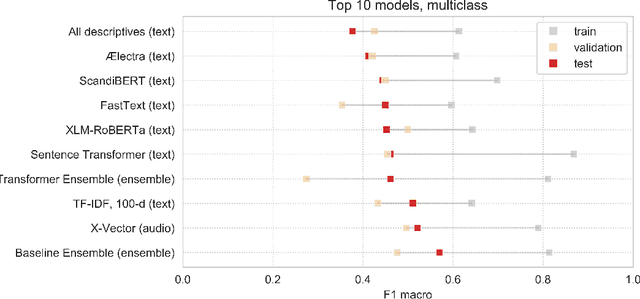
Abstract:Speech patterns have been identified as potential diagnostic markers for neuropsychiatric conditions. However, most studies only compare a single clinical group to healthy controls, whereas clinical practice often requires differentiating between multiple potential diagnoses (multiclass settings). To address this, we assembled a dataset of repeated recordings from 420 participants (67 with major depressive disorder, 106 with schizophrenia and 46 with autism, as well as matched controls), and tested the performance of a range of conventional machine learning models and advanced Transformer models on both binary and multiclass classification, based on voice and text features. While binary models performed comparably to previous research (F1 scores between 0.54-0.75 for autism spectrum disorder, ASD; 0.67-0.92 for major depressive disorder, MDD; and 0.71-0.83 for schizophrenia); when differentiating between multiple diagnostic groups performance decreased markedly (F1 scores between 0.35-0.44 for ASD, 0.57-0.75 for MDD, 0.15-0.66 for schizophrenia, and 0.38-0.52 macro F1). Combining voice and text-based models yielded increased performance, suggesting that they capture complementary diagnostic information. Our results indicate that models trained on binary classification may learn to rely on markers of generic differences between clinical and non-clinical populations, or markers of clinical features that overlap across conditions, rather than identifying markers specific to individual conditions. We provide recommendations for future research in the field, suggesting increased focus on developing larger transdiagnostic datasets that include more fine-grained clinical features, and that can support the development of models that better capture the complexity of neuropsychiatric conditions and naturalistic diagnostic assessment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge