Pingkun Yan
X-WIN: Building Chest Radiograph World Model via Predictive Sensing
Nov 18, 2025Abstract:Chest X-ray radiography (CXR) is an essential medical imaging technique for disease diagnosis. However, as 2D projectional images, CXRs are limited by structural superposition and hence fail to capture 3D anatomies. This limitation makes representation learning and disease diagnosis challenging. To address this challenge, we propose a novel CXR world model named X-WIN, which distills volumetric knowledge from chest computed tomography (CT) by learning to predict its 2D projections in latent space. The core idea is that a world model with internalized knowledge of 3D anatomical structure can predict CXRs under various transformations in 3D space. During projection prediction, we introduce an affinity-guided contrastive alignment loss that leverages mutual similarities to capture rich, correlated information across projections from the same volume. To improve model adaptability, we incorporate real CXRs into training through masked image modeling and employ a domain classifier to encourage statistically similar representations for real and simulated CXRs. Comprehensive experiments show that X-WIN outperforms existing foundation models on diverse downstream tasks using linear probing and few-shot fine-tuning. X-WIN also demonstrates the ability to render 2D projections for reconstructing a 3D CT volume.
OncoReg: Medical Image Registration for Oncological Challenges
Apr 01, 2025Abstract:In modern cancer research, the vast volume of medical data generated is often underutilised due to challenges related to patient privacy. The OncoReg Challenge addresses this issue by enabling researchers to develop and validate image registration methods through a two-phase framework that ensures patient privacy while fostering the development of more generalisable AI models. Phase one involves working with a publicly available dataset, while phase two focuses on training models on a private dataset within secure hospital networks. OncoReg builds upon the foundation established by the Learn2Reg Challenge by incorporating the registration of interventional cone-beam computed tomography (CBCT) with standard planning fan-beam CT (FBCT) images in radiotherapy. Accurate image registration is crucial in oncology, particularly for dynamic treatment adjustments in image-guided radiotherapy, where precise alignment is necessary to minimise radiation exposure to healthy tissues while effectively targeting tumours. This work details the methodology and data behind the OncoReg Challenge and provides a comprehensive analysis of the competition entries and results. Findings reveal that feature extraction plays a pivotal role in this registration task. A new method emerging from this challenge demonstrated its versatility, while established approaches continue to perform comparably to newer techniques. Both deep learning and classical approaches still play significant roles in image registration, with the combination of methods - particularly in feature extraction - proving most effective.
Chest X-ray Foundation Model with Global and Local Representations Integration
Feb 07, 2025Abstract:Chest X-ray (CXR) is the most frequently ordered imaging test, supporting diverse clinical tasks from thoracic disease detection to postoperative monitoring. However, task-specific classification models are limited in scope, require costly labeled data, and lack generalizability to out-of-distribution datasets. To address these challenges, we introduce CheXFound, a self-supervised vision foundation model that learns robust CXR representations and generalizes effectively across a wide range of downstream tasks. We pretrain CheXFound on a curated CXR-1M dataset, comprising over one million unique CXRs from publicly available sources. We propose a Global and Local Representations Integration (GLoRI) module for downstream adaptations, by incorporating disease-specific local features with global image features for enhanced performance in multilabel classification. Our experimental results show that CheXFound outperforms state-of-the-art models in classifying 40 disease findings across different prevalence levels on the CXR-LT 24 dataset and exhibits superior label efficiency on downstream tasks with limited training data. Additionally, CheXFound achieved significant improvements on new tasks with out-of-distribution datasets, including opportunistic cardiovascular disease risk estimation and mortality prediction. These results highlight CheXFound's strong generalization capabilities, enabling diverse adaptations with improved label efficiency. The project source code is publicly available at https://github.com/RPIDIAL/CheXFound.
Evaluating Automated Radiology Report Quality through Fine-Grained Phrasal Grounding of Clinical Findings
Dec 02, 2024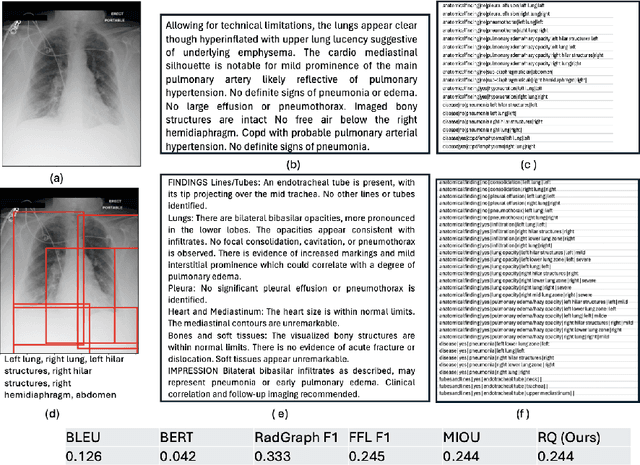
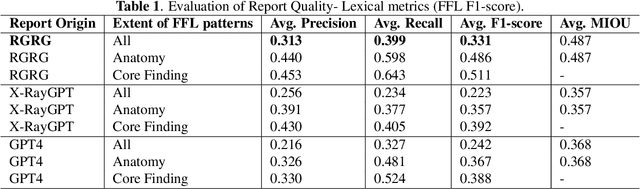
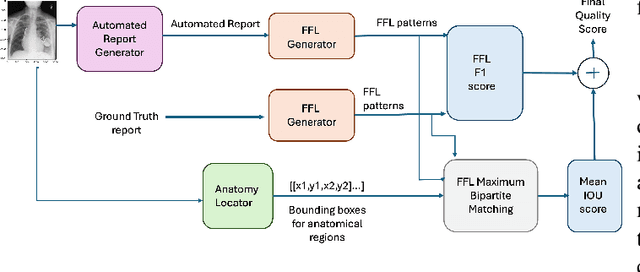

Abstract:Several evaluation metrics have been developed recently to automatically assess the quality of generative AI reports for chest radiographs based only on textual information using lexical, semantic, or clinical named entity recognition methods. In this paper, we develop a new method of report quality evaluation by first extracting fine-grained finding patterns capturing the location, laterality, and severity of a large number of clinical findings. We then performed phrasal grounding to localize their associated anatomical regions on chest radiograph images. The textual and visual measures are then combined to rate the quality of the generated reports. We present results that compare this evaluation metric with other textual metrics on a gold standard dataset derived from the MIMIC collection and show its robustness and sensitivity to factual errors.
Integrating AI in College Education: Positive yet Mixed Experiences with ChatGPT
Jul 08, 2024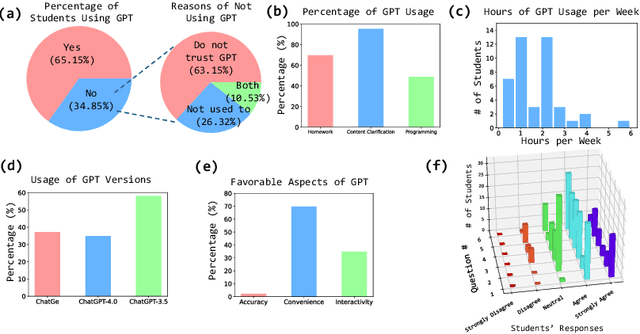
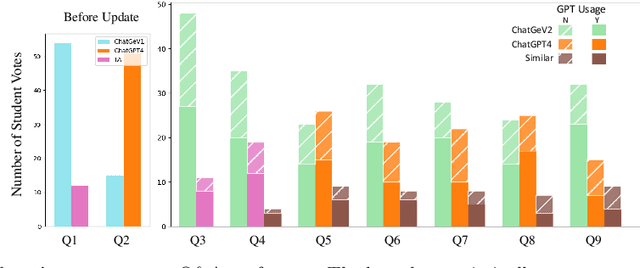
Abstract:The integration of artificial intelligence (AI) chatbots into higher education marks a shift towards a new generation of pedagogical tools, mirroring the arrival of milestones like the internet. With the launch of ChatGPT-4 Turbo in November 2023, we developed a ChatGPT-based teaching application (https://chat.openai.com/g/g-1imx1py4K-chatge-medical-imaging) and integrated it into our undergraduate medical imaging course in the Spring 2024 semester. This study investigates the use of ChatGPT throughout a semester-long trial, providing insights into students' engagement, perception, and the overall educational effectiveness of the technology. We systematically collected and analyzed data concerning students' interaction with ChatGPT, focusing on their attitudes, concerns, and usage patterns. The findings indicate that ChatGPT offers significant advantages such as improved information access and increased interactivity, but its adoption is accompanied by concerns about the accuracy of the information provided and the necessity for well-defined guidelines to optimize its use.
Explaining Chest X-ray Pathology Models using Textual Concepts
Jun 30, 2024


Abstract:Deep learning models have revolutionized medical imaging and diagnostics, yet their opaque nature poses challenges for clinical adoption and trust. Amongst approaches to improve model interpretability, concept-based explanations aim to provide concise and human understandable explanations of any arbitrary classifier. However, such methods usually require a large amount of manually collected data with concept annotation, which is often scarce in the medical domain. In this paper, we propose Conceptual Counterfactual Explanations for Chest X-ray (CoCoX) that leverage existing vision-language models (VLM) joint embedding space to explain black-box classifier outcomes without the need for annotated datasets. Specifically, we utilize textual concepts derived from chest radiography reports and a pre-trained chest radiography-based VLM to explain three common cardiothoracic pathologies. We demonstrate that the explanations generated by our method are semantically meaningful and faithful to underlying pathologies.
Cardiovascular Disease Detection from Multi-View Chest X-rays with BI-Mamba
May 28, 2024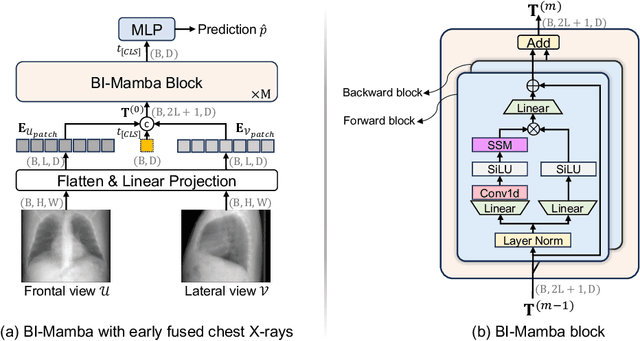
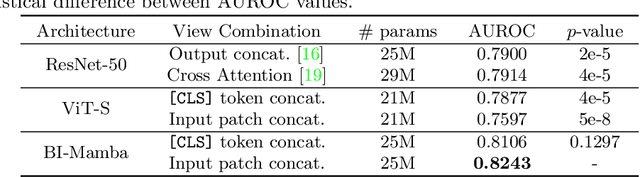
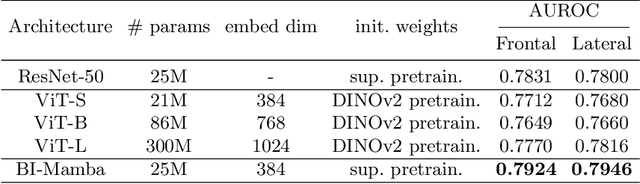
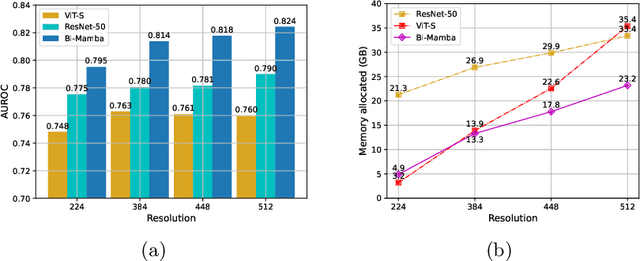
Abstract:Accurate prediction of Cardiovascular disease (CVD) risk in medical imaging is central to effective patient health management. Previous studies have demonstrated that imaging features in computed tomography (CT) can help predict CVD risk. However, CT entails notable radiation exposure, which may result in adverse health effects for patients. In contrast, chest X-ray emits significantly lower levels of radiation, offering a safer option. This rationale motivates our investigation into the feasibility of using chest X-ray for predicting CVD risk. Convolutional Neural Networks (CNNs) and Transformers are two established network architectures for computer-aided diagnosis. However, they struggle to model very high resolution chest X-ray due to the lack of large context modeling power or quadratic time complexity. Inspired by state space sequence models (SSMs), a new class of network architectures with competitive sequence modeling power as Transfomers and linear time complexity, we propose Bidirectional Image Mamba (BI-Mamba) to complement the unidirectional SSMs with opposite directional information. BI-Mamba utilizes parallel forward and backwark blocks to encode longe-range dependencies of multi-view chest X-rays. We conduct extensive experiments on images from 10,395 subjects in National Lung Screening Trail (NLST). Results show that BI-Mamba outperforms ResNet-50 and ViT-S with comparable parameter size, and saves significant amount of GPU memory during training. Besides, BI-Mamba achieves promising performance compared with previous state of the art in CT, unraveling the potential of chest X-ray for CVD risk prediction.
Disease-informed Adaptation of Vision-Language Models
May 24, 2024Abstract:In medical image analysis, the expertise scarcity and the high cost of data annotation limits the development of large artificial intelligence models. This paper investigates the potential of transfer learning with pre-trained vision-language models (VLMs) in this domain. Currently, VLMs still struggle to transfer to the underrepresented diseases with minimal presence and new diseases entirely absent from the pretraining dataset. We argue that effective adaptation of VLMs hinges on the nuanced representation learning of disease concepts. By capitalizing on the joint visual-linguistic capabilities of VLMs, we introduce disease-informed contextual prompting in a novel disease prototype learning framework. This approach enables VLMs to grasp the concepts of new disease effectively and efficiently, even with limited data. Extensive experiments across multiple image modalities showcase notable enhancements in performance compared to existing techniques.
General Purpose Image Encoder DINOv2 for Medical Image Registration
Feb 24, 2024



Abstract:Existing medical image registration algorithms rely on either dataset specific training or local texture-based features to align images. The former cannot be reliably implemented without large modality-specific training datasets, while the latter lacks global semantics thus could be easily trapped at local minima. In this paper, we present a training-free deformable image registration method, DINO-Reg, leveraging a general purpose image encoder DINOv2 for image feature extraction. The DINOv2 encoder was trained using the ImageNet data containing natural images. We used the pretrained DINOv2 without any finetuning. Our method feeds the DINOv2 encoded features into a discrete optimizer to find the optimal deformable registration field. We conducted a series of experiments to understand the behavior and role of such a general purpose image encoder in the application of image registration. Combined with handcrafted features, our method won the first place in the recent OncoReg Challenge. To our knowledge, this is the first application of general vision foundation models in medical image registration.
Multimodal Neurodegenerative Disease Subtyping Explained by ChatGPT
Jan 31, 2024Abstract:Alzheimer's disease (AD) is the most prevalent neurodegenerative disease; yet its currently available treatments are limited to stopping disease progression. Moreover, effectiveness of these treatments is not guaranteed due to the heterogenetiy of the disease. Therefore, it is essential to be able to identify the disease subtypes at a very early stage. Current data driven approaches are able to classify the subtypes at later stages of AD or related disorders, but struggle when predicting at the asymptomatic or prodromal stage. Moreover, most existing models either lack explainability behind the classification or only use a single modality for the assessment, limiting scope of its analysis. Thus, we propose a multimodal framework that uses early-stage indicators such as imaging, genetics and clinical assessments to classify AD patients into subtypes at early stages. Similarly, we build prompts and use large language models, such as ChatGPT, to interpret the findings of our model. In our framework, we propose a tri-modal co-attention mechanism (Tri-COAT) to explicitly learn the cross-modal feature associations. Our proposed model outperforms baseline models and provides insight into key cross-modal feature associations supported by known biological mechanisms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge