Parisa Kaviani
Evaluating Automated Radiology Report Quality through Fine-Grained Phrasal Grounding of Clinical Findings
Dec 02, 2024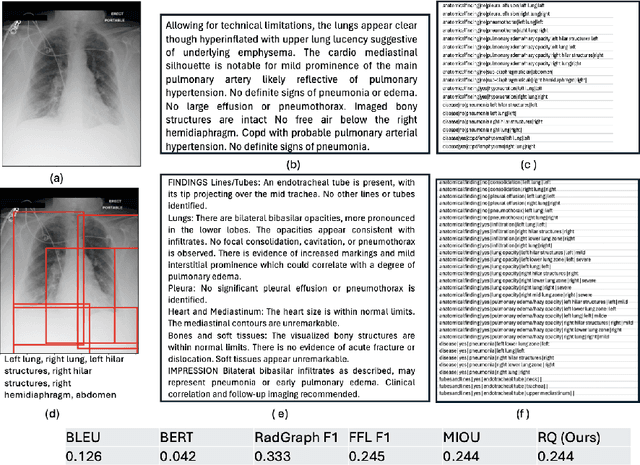
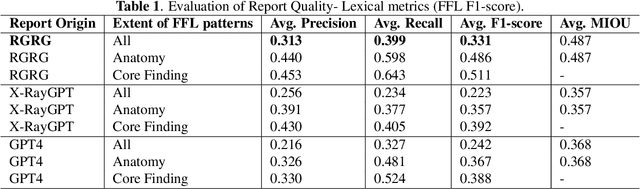
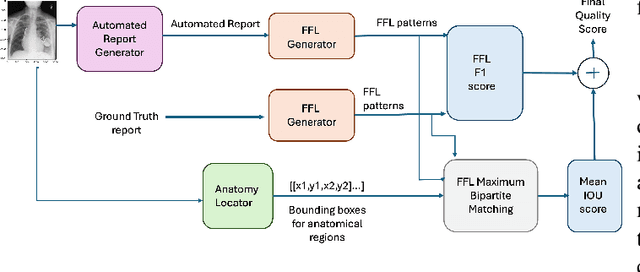

Abstract:Several evaluation metrics have been developed recently to automatically assess the quality of generative AI reports for chest radiographs based only on textual information using lexical, semantic, or clinical named entity recognition methods. In this paper, we develop a new method of report quality evaluation by first extracting fine-grained finding patterns capturing the location, laterality, and severity of a large number of clinical findings. We then performed phrasal grounding to localize their associated anatomical regions on chest radiograph images. The textual and visual measures are then combined to rate the quality of the generated reports. We present results that compare this evaluation metric with other textual metrics on a gold standard dataset derived from the MIMIC collection and show its robustness and sensitivity to factual errors.
Cross-Institutional Structured Radiology Reporting for Lung Cancer Screening Using a Dynamic Template-Constrained Large Language Model
Sep 26, 2024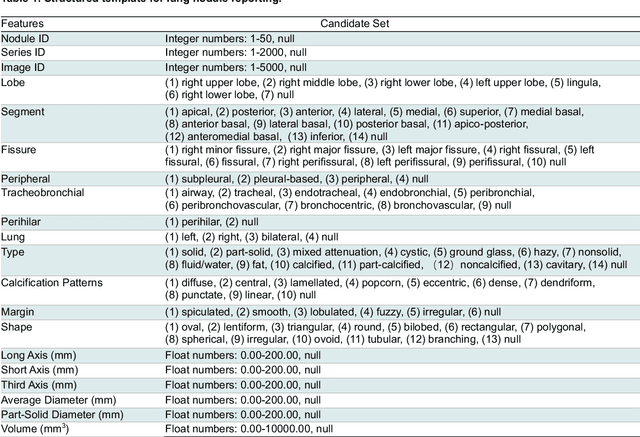
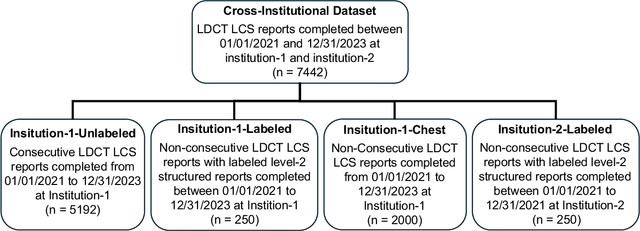
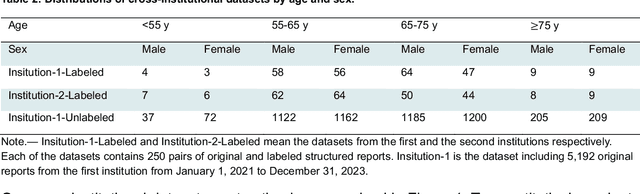
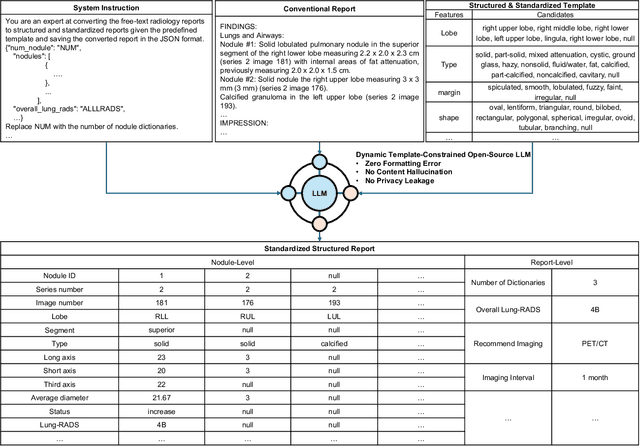
Abstract:Structured radiology reporting is advantageous for optimizing clinical workflows and patient outcomes. Current LLMs in creating structured reports face the challenges of formatting errors, content hallucinations, and privacy leakage concerns when uploaded to external servers. We aim to develop an enhanced open-source LLM for creating structured and standardized LCS reports from free-text descriptions. After institutional IRB approvals, 5,442 de-identified LCS reports from two institutions were retrospectively analyzed. 500 reports were randomly selected from the two institutions evenly and then manually labeled for evaluation. Two radiologists from the two institutions developed a standardized template including 29 features for lung nodule reporting. We proposed template-constrained decoding to enhance state-of-the-art open-source LLMs, including LLAMA, Qwen, and Mistral. The LLM performance was extensively evaluated in terms of F1 score, confidence interval, McNemar test, and z-test. Based on the structured reports created from the large-scale dataset, a nodule-level retrieval system was prototyped and an automatic statistical analysis was performed. Our software, vLLM-structure, is publicly available for local deployment with enhanced LLMs. Our template-constrained decoding approach consistently enhanced the LLM performance on multi-institutional datasets, with neither formatting errors nor content hallucinations. Our method improved the best open-source LLAMA-3.1 405B by up to 10.42%, and outperformed GPT-4o by 17.19%. A novel nodule retrieval system was successfully prototyped and demonstrated on a large-scale multimodal database using our enhanced LLM technologies. The automatically derived statistical distributions were closely consistent with the prior findings in terms of nodule type, location, size, status, and Lung-RADS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge