Christopher T. Whitlow
MCR-VQGAN: A Scalable and Cost-Effective Tau PET Synthesis Approach for Alzheimer's Disease Imaging
Dec 17, 2025



Abstract:Tau positron emission tomography (PET) is a critical diagnostic modality for Alzheimer's disease (AD) because it visualizes and quantifies neurofibrillary tangles, a hallmark of AD pathology. However, its widespread clinical adoption is hindered by significant challenges, such as radiation exposure, limited availability, high clinical workload, and substantial financial costs. To overcome these limitations, we propose Multi-scale CBAM Residual Vector Quantized Generative Adversarial Network (MCR-VQGAN) to synthesize high-fidelity tau PET images from structural T1-weighted MRI scans. MCR-VQGAN improves standard VQGAN by integrating three key architectural enhancements: multi-scale convolutions, ResNet blocks, and Convolutional Block Attention Modules (CBAM). Using 222 paired structural T1-weighted MRI and tau PET scans from Alzheimer's Disease Neuroimaging Initiative (ADNI), we trained and compared MCR-VQGAN with cGAN, WGAN-GP, CycleGAN, and VQGAN. Our proposed model achieved superior image synthesis performance across all metrics: MSE of 0.0056 +/- 0.0061, PSNR of 24.39 +/- 4.49 dB, and SSIM of 0.9000 +/- 0.0453. To assess the clinical utility of the synthetic images, we trained and evaluated a CNN-based AD classifier. The classifier achieved comparable accuracy when tested on real (63.64%) and synthetic (65.91%) images. This result indicates that our synthesis process successfully preserves diagnostically relevant features without significant information loss. Our results demonstrate that MCR-VQGAN can offer a reliable and scalable surrogate for conventional tau PET imaging, potentially improving the accessibility and scalability of tau imaging biomarkers for AD research and clinical workflows.
Nonhuman Primate Brain Tissue Segmentation Using a Transfer Learning Approach
Apr 01, 2025Abstract:Non-human primates (NHPs) serve as critical models for understanding human brain function and neurological disorders due to their close evolutionary relationship with humans. Accurate brain tissue segmentation in NHPs is critical for understanding neurological disorders, but challenging due to the scarcity of annotated NHP brain MRI datasets, the small size of the NHP brain, the limited resolution of available imaging data and the anatomical differences between human and NHP brains. To address these challenges, we propose a novel approach utilizing STU-Net with transfer learning to leverage knowledge transferred from human brain MRI data to enhance segmentation accuracy in the NHP brain MRI, particularly when training data is limited. The combination of STU-Net and transfer learning effectively delineates complex tissue boundaries and captures fine anatomical details specific to NHP brains. Notably, our method demonstrated improvement in segmenting small subcortical structures such as putamen and thalamus that are challenging to resolve with limited spatial resolution and tissue contrast, and achieved DSC of over 0.88, IoU over 0.8 and HD95 under 7. This study introduces a robust method for multi-class brain tissue segmentation in NHPs, potentially accelerating research in evolutionary neuroscience and preclinical studies of neurological disorders relevant to human health.
Cross-Institutional Structured Radiology Reporting for Lung Cancer Screening Using a Dynamic Template-Constrained Large Language Model
Sep 26, 2024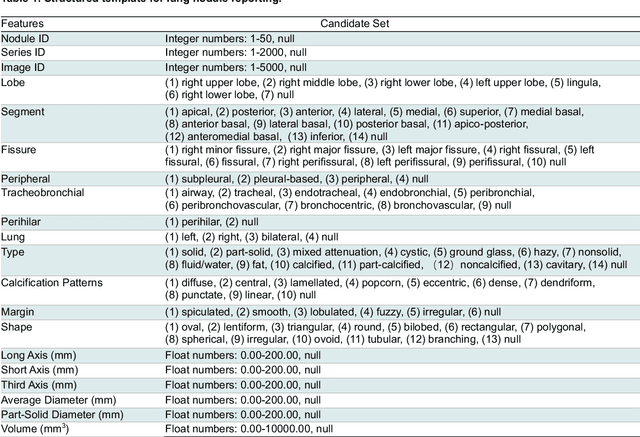
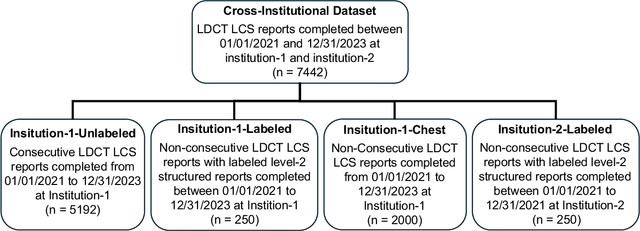
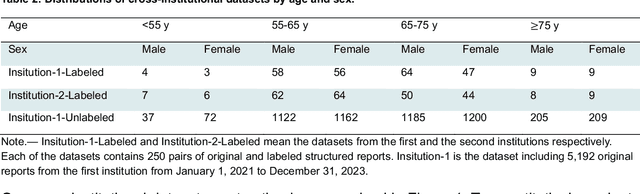
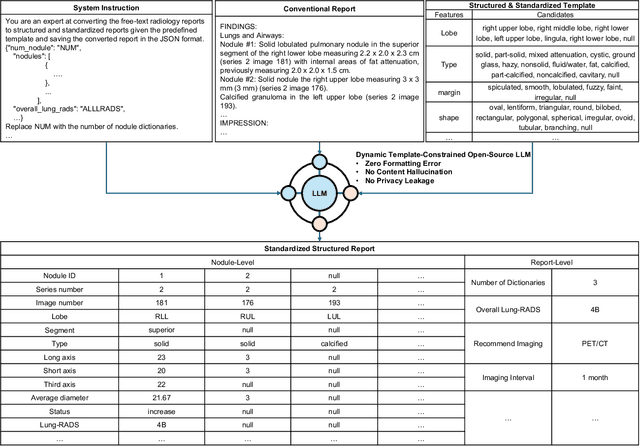
Abstract:Structured radiology reporting is advantageous for optimizing clinical workflows and patient outcomes. Current LLMs in creating structured reports face the challenges of formatting errors, content hallucinations, and privacy leakage concerns when uploaded to external servers. We aim to develop an enhanced open-source LLM for creating structured and standardized LCS reports from free-text descriptions. After institutional IRB approvals, 5,442 de-identified LCS reports from two institutions were retrospectively analyzed. 500 reports were randomly selected from the two institutions evenly and then manually labeled for evaluation. Two radiologists from the two institutions developed a standardized template including 29 features for lung nodule reporting. We proposed template-constrained decoding to enhance state-of-the-art open-source LLMs, including LLAMA, Qwen, and Mistral. The LLM performance was extensively evaluated in terms of F1 score, confidence interval, McNemar test, and z-test. Based on the structured reports created from the large-scale dataset, a nodule-level retrieval system was prototyped and an automatic statistical analysis was performed. Our software, vLLM-structure, is publicly available for local deployment with enhanced LLMs. Our template-constrained decoding approach consistently enhanced the LLM performance on multi-institutional datasets, with neither formatting errors nor content hallucinations. Our method improved the best open-source LLAMA-3.1 405B by up to 10.42%, and outperformed GPT-4o by 17.19%. A novel nodule retrieval system was successfully prototyped and demonstrated on a large-scale multimodal database using our enhanced LLM technologies. The automatically derived statistical distributions were closely consistent with the prior findings in terms of nodule type, location, size, status, and Lung-RADS.
Head-Neck Dual-energy CT Contrast Media Reduction Using Diffusion Models
Aug 24, 2023



Abstract:Iodinated contrast media is essential for dual-energy computed tomography (DECT) angiography. Previous studies show that iodinated contrast media may cause side effects, and the interruption of the supply chain in 2022 led to a severe contrast media shortage in the US. Both factors justify the necessity of contrast media reduction in relevant clinical applications. In this study, we propose a diffusion model-based deep learning framework to address this challenge. First, we simulate different levels of low contrast dosage DECT scans from the standard normal contrast dosage DECT scans using material decomposition. Conditional denoising diffusion probabilistic models are then trained to enhance the contrast media and create contrast-enhanced images. Our results demonstrate that the proposed methods can generate high-quality contrast-enhanced results even for images obtained with as low as 12.5% of the normal contrast dosage. Furthermore, our method outperforms selected competing methods in a human reader study.
Translating Radiology Reports into Plain Language using ChatGPT and GPT-4 with Prompt Learning: Promising Results, Limitations, and Potential
Mar 29, 2023



Abstract:The large language model called ChatGPT has drawn extensively attention because of its human-like expression and reasoning abilities. In this study, we investigate the feasibility of using ChatGPT in experiments on using ChatGPT to translate radiology reports into plain language for patients and healthcare providers so that they are educated for improved healthcare. Radiology reports from 62 low-dose chest CT lung cancer screening scans and 76 brain MRI metastases screening scans were collected in the first half of February for this study. According to the evaluation by radiologists, ChatGPT can successfully translate radiology reports into plain language with an average score of 4.27 in the five-point system with 0.08 places of information missing and 0.07 places of misinformation. In terms of the suggestions provided by ChatGPT, they are general relevant such as keeping following-up with doctors and closely monitoring any symptoms, and for about 37% of 138 cases in total ChatGPT offers specific suggestions based on findings in the report. ChatGPT also presents some randomness in its responses with occasionally over-simplified or neglected information, which can be mitigated using a more detailed prompt. Furthermore, ChatGPT results are compared with a newly released large model GPT-4, showing that GPT-4 can significantly improve the quality of translated reports. Our results show that it is feasible to utilize large language models in clinical education, and further efforts are needed to address limitations and maximize their potential.
SoftDropConnect (SDC) -- Effective and Efficient Quantification of the Network Uncertainty in Deep MR Image Analysis
Jan 20, 2022



Abstract:Recently, deep learning has achieved remarkable successes in medical image analysis. Although deep neural networks generate clinically important predictions, they have inherent uncertainty. Such uncertainty is a major barrier to report these predictions with confidence. In this paper, we propose a novel yet simple Bayesian inference approach called SoftDropConnect (SDC) to quantify the network uncertainty in medical imaging tasks with gliomas segmentation and metastases classification as initial examples. Our key idea is that during training and testing SDC modulates network parameters continuously so as to allow affected information processing channels still in operation, instead of disabling them as Dropout or DropConnet does. When compared with three popular Bayesian inference methods including Bayes By Backprop, Dropout, and DropConnect, our SDC method (SDC-W after optimization) outperforms the three competing methods with a substantial margin. Quantitatively, our proposed method generates results withsubstantially improved prediction accuracy (by 10.0%, 5.4% and 3.7% respectively for segmentation in terms of dice score; by 11.7%, 3.9%, 8.7% on classification in terms of test accuracy) and greatly reduced uncertainty in terms of mutual information (by 64%, 33% and 70% on segmentation; 98%, 88%, and 88% on classification). Our approach promises to deliver better diagnostic performance and make medical AI imaging more explainable and trustworthy.
A transformer-based deep learning approach for classifying brain metastases into primary organ sites using clinical whole brain MRI images
Oct 07, 2021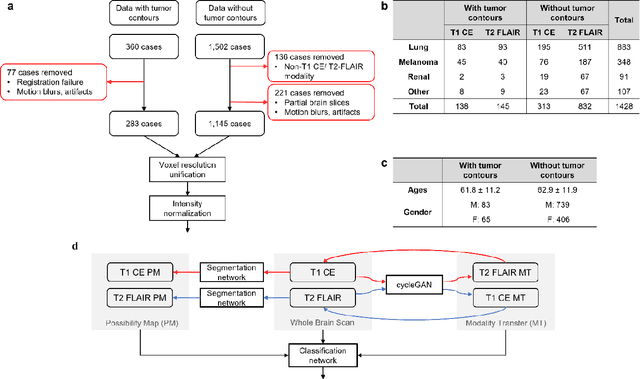
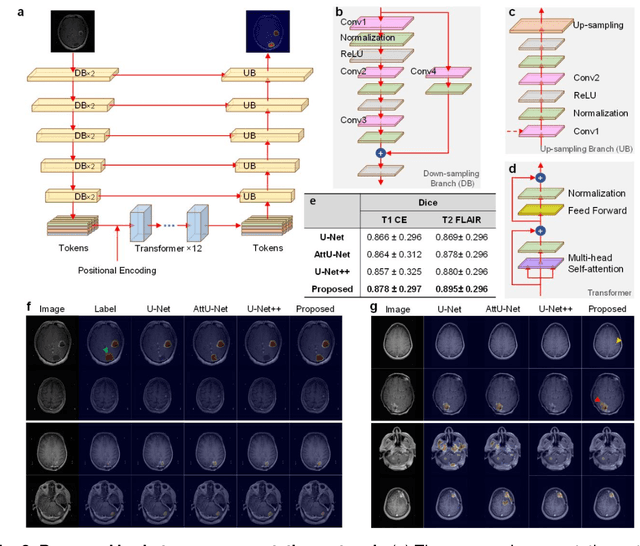
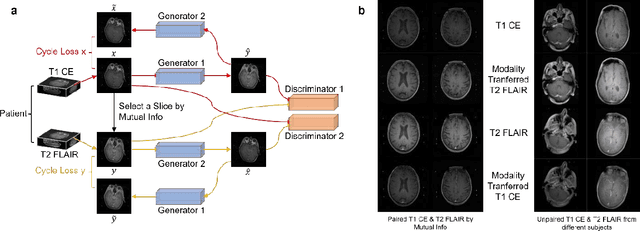
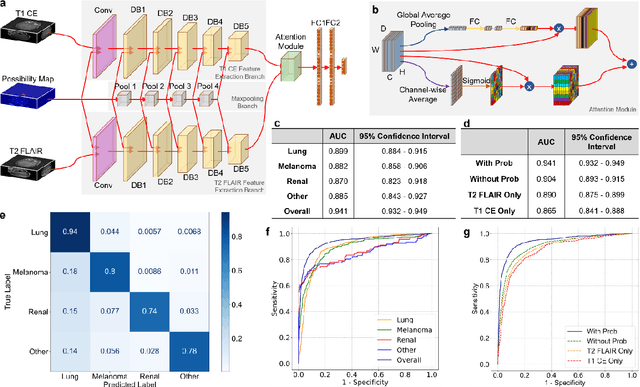
Abstract:The treatment decisions for brain metastatic disease are driven by knowledge of the primary organ site cancer histology, often requiring invasive biopsy. This study aims to develop a novel deep learning approach for accurate and rapid non-invasive identification of brain metastatic tumor histology with conventional whole-brain MRI. The use of clinical whole-brain data and the end-to-end pipeline obviate external human intervention. This IRB-approved single-site retrospective study was comprised of patients (n=1,293) referred for MRI treatment-planning and gamma knife radiosurgery from July 2000 to May 2019. Contrast-enhanced T1-weighted contrast enhanced and T2-weighted-Fluid-Attenuated Inversion Recovery brain MRI exams (n=1,428) were minimally preprocessed (voxel resolution unification and signal-intensity rescaling/normalization), requiring only seconds per an MRI scan, and input into the proposed deep learning workflow for tumor segmentation, modality transfer, and primary site classification associated with brain metastatic disease in one of four classes (lung, melanoma, renal, and other). Ten-fold cross-validation generated the overall AUC of 0.941, lung class AUC of 0.899, melanoma class AUC of 0.882, renal class AUC of 0.870, and other class AUC of 0.885. It is convincingly established that whole-brain imaging features would be sufficiently discriminative to allow accurate diagnosis of the primary organ site of malignancy. Our end-to-end deep learning-based radiomic method has a great translational potential for classifying metastatic tumor types using whole-brain MRI images, without additional human intervention. Further refinement may offer invaluable tools to expedite primary organ site cancer identification for treatment of brain metastatic disease and improvement of patient outcomes and survival.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge