Metin N. Gurcan
A transformer-based deep learning approach for classifying brain metastases into primary organ sites using clinical whole brain MRI images
Oct 07, 2021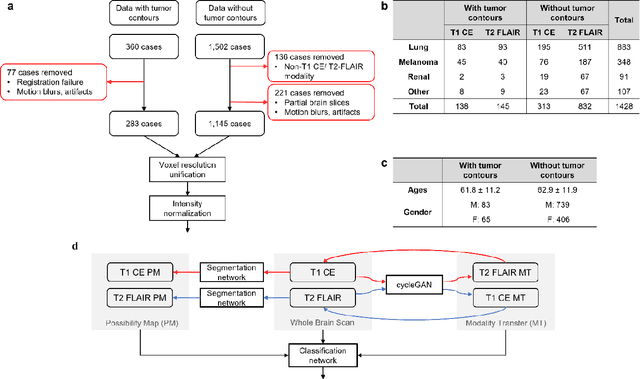
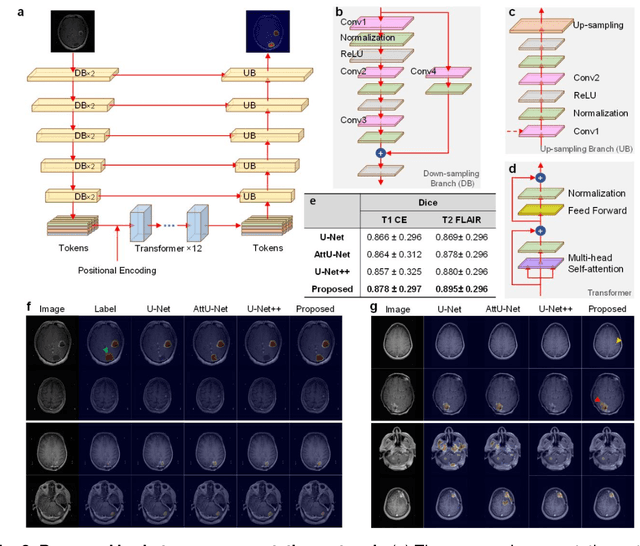
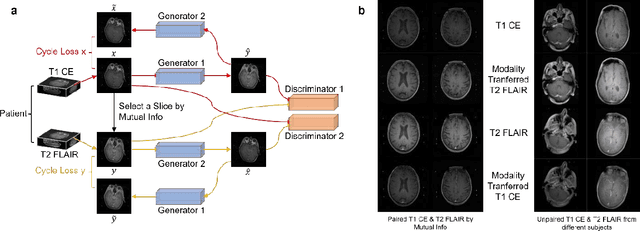
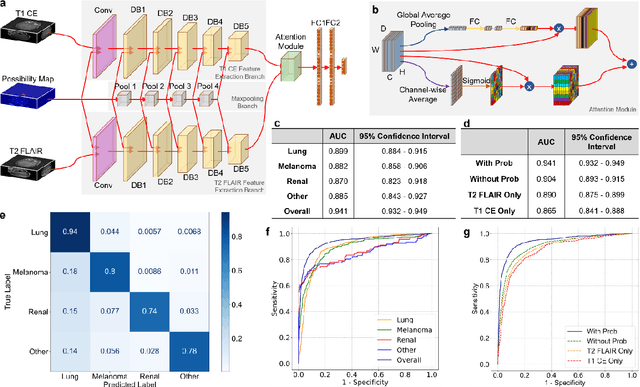
Abstract:The treatment decisions for brain metastatic disease are driven by knowledge of the primary organ site cancer histology, often requiring invasive biopsy. This study aims to develop a novel deep learning approach for accurate and rapid non-invasive identification of brain metastatic tumor histology with conventional whole-brain MRI. The use of clinical whole-brain data and the end-to-end pipeline obviate external human intervention. This IRB-approved single-site retrospective study was comprised of patients (n=1,293) referred for MRI treatment-planning and gamma knife radiosurgery from July 2000 to May 2019. Contrast-enhanced T1-weighted contrast enhanced and T2-weighted-Fluid-Attenuated Inversion Recovery brain MRI exams (n=1,428) were minimally preprocessed (voxel resolution unification and signal-intensity rescaling/normalization), requiring only seconds per an MRI scan, and input into the proposed deep learning workflow for tumor segmentation, modality transfer, and primary site classification associated with brain metastatic disease in one of four classes (lung, melanoma, renal, and other). Ten-fold cross-validation generated the overall AUC of 0.941, lung class AUC of 0.899, melanoma class AUC of 0.882, renal class AUC of 0.870, and other class AUC of 0.885. It is convincingly established that whole-brain imaging features would be sufficiently discriminative to allow accurate diagnosis of the primary organ site of malignancy. Our end-to-end deep learning-based radiomic method has a great translational potential for classifying metastatic tumor types using whole-brain MRI images, without additional human intervention. Further refinement may offer invaluable tools to expedite primary organ site cancer identification for treatment of brain metastatic disease and improvement of patient outcomes and survival.
Radiopathomics: Integration of radiographic and histologic characteristics for prognostication in glioblastoma
Sep 19, 2019



Abstract:Both radiographic (Rad) imaging, such as multi-parametric magnetic resonance imaging, and digital pathology (Path) images captured from tissue samples are currently acquired as standard clinical practice for glioblastoma tumors. Both these data streams have been separately used for diagnosis and treatment planning, despite the fact that they provide complementary information. In this research work, we aimed to assess the potential of both Rad and Path images in combination and comparison. An extensive set of engineered features was extracted from delineated tumor regions in Rad images, comprising T1, T1-Gd, T2, T2-FLAIR, and 100 random patches extracted from Path images. Specifically, the features comprised descriptors of intensity, histogram, and texture, mainly quantified via gray-level-co-occurrence matrix and gray-level-run-length matrices. Features extracted from images of 107 glioblastoma patients, downloaded from The Cancer Imaging Archive, were run through support vector machine for classification using leave-one-out cross-validation mechanism, and through support vector regression for prediction of continuous survival outcome. The Pearson correlation coefficient was estimated to be 0.75, 0.74, and 0.78 for Rad, Path and RadPath data. The area-under the receiver operating characteristic curve was estimated to be 0.74, 0.76 and 0.80 for Rad, Path and RadPath data, when patients were discretized into long- and short-survival groups based on average survival cutoff. Our results support the notion that synergistically using Rad and Path images may lead to better prognosis at the initial presentation of the disease, thereby facilitating the targeted enrollment of patients into clinical trials.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge