Michael D. Chan
A transformer-based deep learning approach for classifying brain metastases into primary organ sites using clinical whole brain MRI images
Oct 07, 2021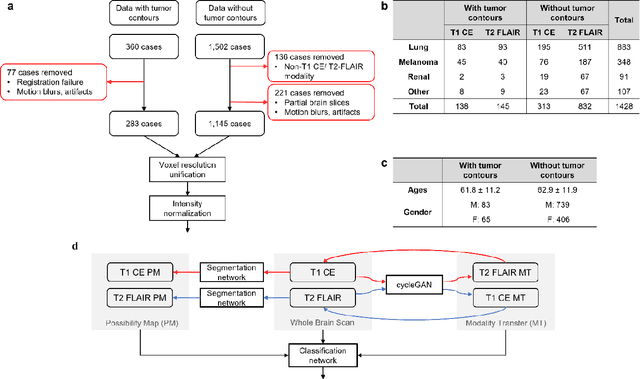
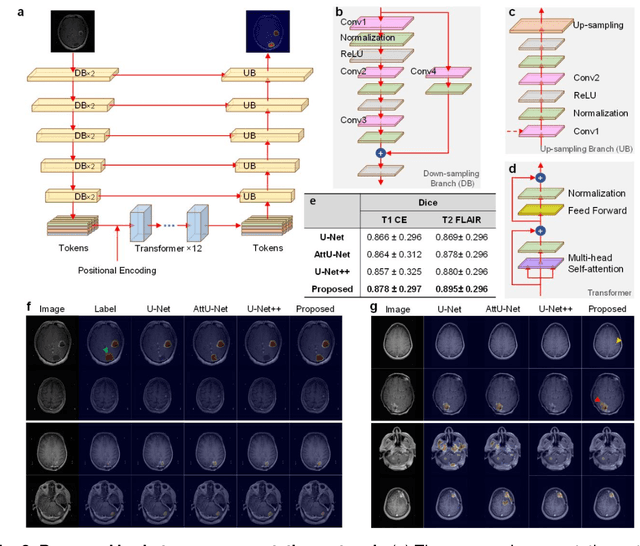
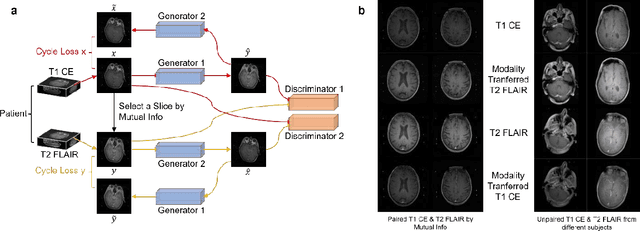
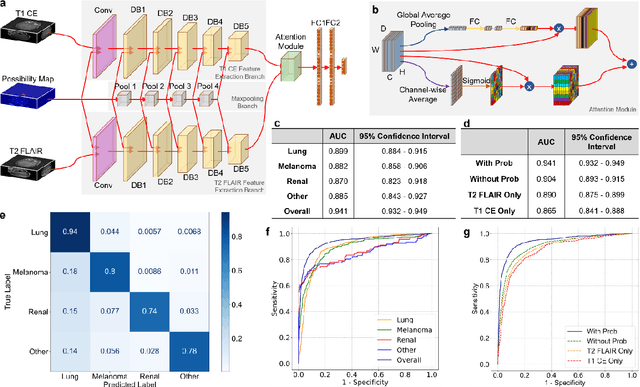
Abstract:The treatment decisions for brain metastatic disease are driven by knowledge of the primary organ site cancer histology, often requiring invasive biopsy. This study aims to develop a novel deep learning approach for accurate and rapid non-invasive identification of brain metastatic tumor histology with conventional whole-brain MRI. The use of clinical whole-brain data and the end-to-end pipeline obviate external human intervention. This IRB-approved single-site retrospective study was comprised of patients (n=1,293) referred for MRI treatment-planning and gamma knife radiosurgery from July 2000 to May 2019. Contrast-enhanced T1-weighted contrast enhanced and T2-weighted-Fluid-Attenuated Inversion Recovery brain MRI exams (n=1,428) were minimally preprocessed (voxel resolution unification and signal-intensity rescaling/normalization), requiring only seconds per an MRI scan, and input into the proposed deep learning workflow for tumor segmentation, modality transfer, and primary site classification associated with brain metastatic disease in one of four classes (lung, melanoma, renal, and other). Ten-fold cross-validation generated the overall AUC of 0.941, lung class AUC of 0.899, melanoma class AUC of 0.882, renal class AUC of 0.870, and other class AUC of 0.885. It is convincingly established that whole-brain imaging features would be sufficiently discriminative to allow accurate diagnosis of the primary organ site of malignancy. Our end-to-end deep learning-based radiomic method has a great translational potential for classifying metastatic tumor types using whole-brain MRI images, without additional human intervention. Further refinement may offer invaluable tools to expedite primary organ site cancer identification for treatment of brain metastatic disease and improvement of patient outcomes and survival.
DC-Al GAN: Pseudoprogression and True Tumor Progression of Glioblastoma multiform Image Classification Based On DCGAN and Alexnet
Feb 26, 2019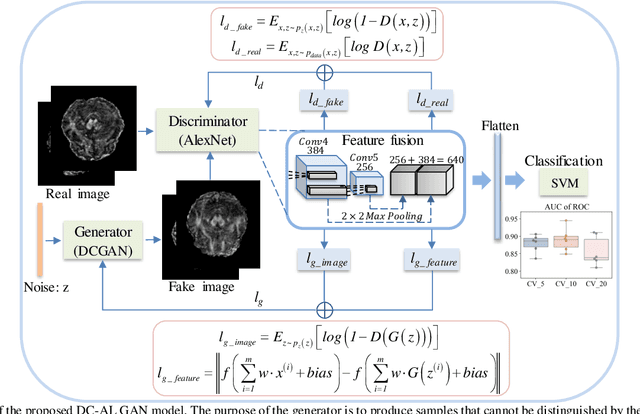
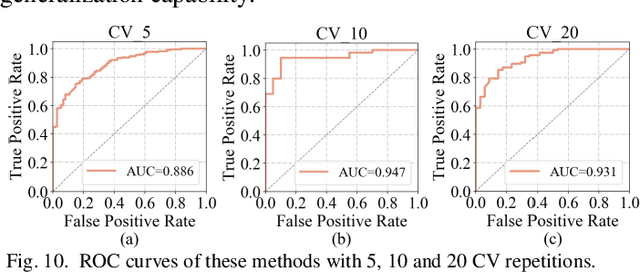
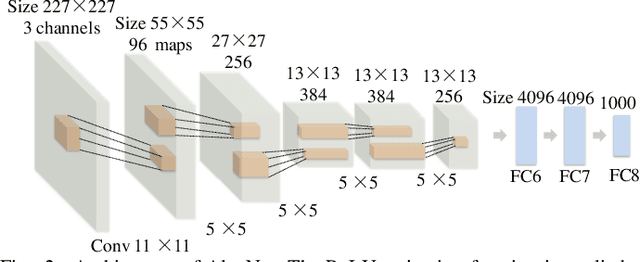
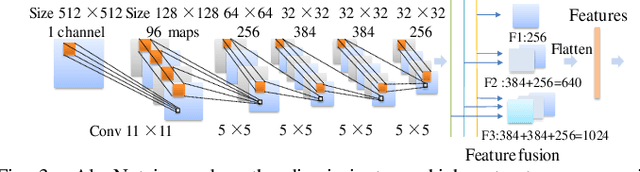
Abstract:Glioblastoma multiform (GBM) is a kind of head tumor with an extraordinarily complex treatment process. The survival period is typically 14-16 months, and the 2 year survival rate is approximately 26%-33%. The clinical treatment strategies for the pseudoprogression (PsP) and true tumor progression (TTP) of GBM are different, so accurately distinguishing these two conditions is particularly significant.As PsP and TTP of GBM are similar in shape and other characteristics, it is hard to distinguish these two forms with precision. In order to differentiate them accurately, this paper introduces a feature learning method based on a generative adversarial network: DC-Al GAN. GAN consists of two architectures: generator and discriminator. Alexnet is used as the discriminator in this work. Owing to the adversarial and competitive relationship between generator and discriminator, the latter extracts highly concise features during training. In DC-Al GAN, features are extracted from Alexnet in the final classification phase, and the highly nature of them contributes positively to the classification accuracy.The generator in DC-Al GAN is modified by the deep convolutional generative adversarial network (DCGAN) by adding three convolutional layers. This effectively generates higher resolution sample images. Feature fusion is used to combine high layer features with low layer features, allowing for the creation and use of more precise features for classification. The experimental results confirm that DC-Al GAN achieves high accuracy on GBM datasets for PsP and TTP image classification, which is superior to other state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge