Xiaobo Zhou
CometNet: Contextual Motif-guided Long-term Time Series Forecasting
Nov 11, 2025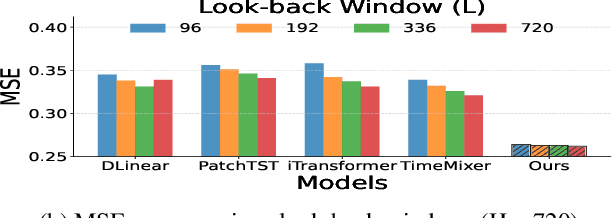
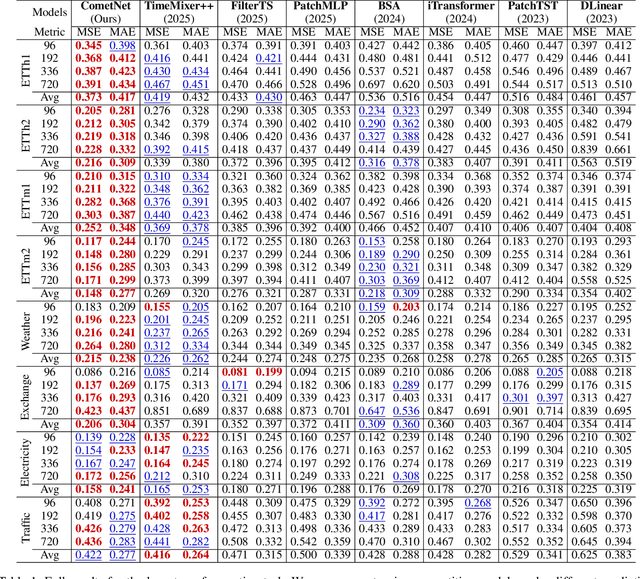
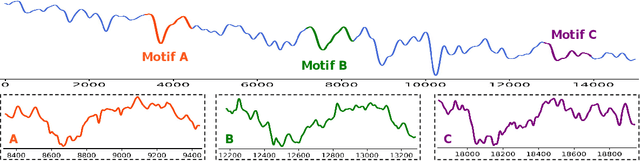
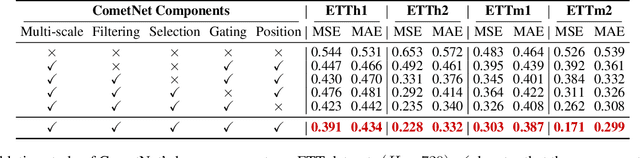
Abstract:Long-term Time Series Forecasting is crucial across numerous critical domains, yet its accuracy remains fundamentally constrained by the receptive field bottleneck in existing models. Mainstream Transformer- and Multi-layer Perceptron (MLP)-based methods mainly rely on finite look-back windows, limiting their ability to model long-term dependencies and hurting forecasting performance. Naively extending the look-back window proves ineffective, as it not only introduces prohibitive computational complexity, but also drowns vital long-term dependencies in historical noise. To address these challenges, we propose CometNet, a novel Contextual Motif-guided Long-term Time Series Forecasting framework. CometNet first introduces a Contextual Motif Extraction module that identifies recurrent, dominant contextual motifs from complex historical sequences, providing extensive temporal dependencies far exceeding limited look-back windows; Subsequently, a Motif-guided Forecasting module is proposed, which integrates the extracted dominant motifs into forecasting. By dynamically mapping the look-back window to its relevant motifs, CometNet effectively harnesses their contextual information to strengthen long-term forecasting capability. Extensive experimental results on eight real-world datasets have demonstrated that CometNet significantly outperforms current state-of-the-art (SOTA) methods, particularly on extended forecast horizons.
StreamSTGS: Streaming Spatial and Temporal Gaussian Grids for Real-Time Free-Viewpoint Video
Nov 08, 2025Abstract:Streaming free-viewpoint video~(FVV) in real-time still faces significant challenges, particularly in training, rendering, and transmission efficiency. Harnessing superior performance of 3D Gaussian Splatting~(3DGS), recent 3DGS-based FVV methods have achieved notable breakthroughs in both training and rendering. However, the storage requirements of these methods can reach up to $10$MB per frame, making stream FVV in real-time impossible. To address this problem, we propose a novel FVV representation, dubbed StreamSTGS, designed for real-time streaming. StreamSTGS represents a dynamic scene using canonical 3D Gaussians, temporal features, and a deformation field. For high compression efficiency, we encode canonical Gaussian attributes as 2D images and temporal features as a video. This design not only enables real-time streaming, but also inherently supports adaptive bitrate control based on network condition without any extra training. Moreover, we propose a sliding window scheme to aggregate adjacent temporal features to learn local motions, and then introduce a transformer-guided auxiliary training module to learn global motions. On diverse FVV benchmarks, StreamSTGS demonstrates competitive performance on all metrics compared to state-of-the-art methods. Notably, StreamSTGS increases the PSNR by an average of $1$dB while reducing the average frame size to just $170$KB. The code is publicly available on https://github.com/kkkzh/StreamSTGS.
SitPose: Real-Time Detection of Sitting Posture and Sedentary Behavior Using Ensemble Learning With Depth Sensor
Dec 16, 2024



Abstract:Poor sitting posture can lead to various work-related musculoskeletal disorders (WMSDs). Office employees spend approximately 81.8% of their working time seated, and sedentary behavior can result in chronic diseases such as cervical spondylosis and cardiovascular diseases. To address these health concerns, we present SitPose, a sitting posture and sedentary detection system utilizing the latest Kinect depth camera. The system tracks 3D coordinates of bone joint points in real-time and calculates the angle values of related joints. We established a dataset containing six different sitting postures and one standing posture, totaling 33,409 data points, by recruiting 36 participants. We applied several state-of-the-art machine learning algorithms to the dataset and compared their performance in recognizing the sitting poses. Our results show that the ensemble learning model based on the soft voting mechanism achieves the highest F1 score of 98.1%. Finally, we deployed the SitPose system based on this ensemble model to encourage better sitting posture and to reduce sedentary habits.
TimeFormer: Capturing Temporal Relationships of Deformable 3D Gaussians for Robust Reconstruction
Nov 18, 2024Abstract:Dynamic scene reconstruction is a long-term challenge in 3D vision. Recent methods extend 3D Gaussian Splatting to dynamic scenes via additional deformation fields and apply explicit constraints like motion flow to guide the deformation. However, they learn motion changes from individual timestamps independently, making it challenging to reconstruct complex scenes, particularly when dealing with violent movement, extreme-shaped geometries, or reflective surfaces. To address the above issue, we design a plug-and-play module called TimeFormer to enable existing deformable 3D Gaussians reconstruction methods with the ability to implicitly model motion patterns from a learning perspective. Specifically, TimeFormer includes a Cross-Temporal Transformer Encoder, which adaptively learns the temporal relationships of deformable 3D Gaussians. Furthermore, we propose a two-stream optimization strategy that transfers the motion knowledge learned from TimeFormer to the base stream during the training phase. This allows us to remove TimeFormer during inference, thereby preserving the original rendering speed. Extensive experiments in the multi-view and monocular dynamic scenes validate qualitative and quantitative improvement brought by TimeFormer. Project Page: https://patrickddj.github.io/TimeFormer/
DS-NeRV: Implicit Neural Video Representation with Decomposed Static and Dynamic Codes
Mar 23, 2024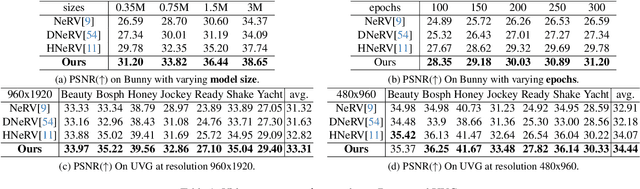
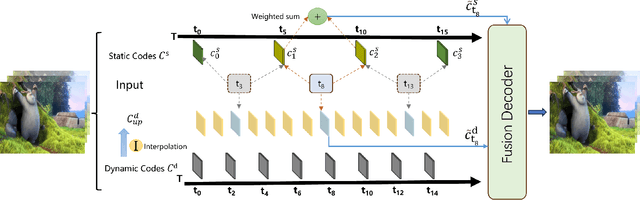
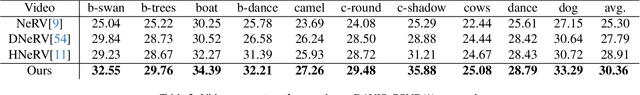
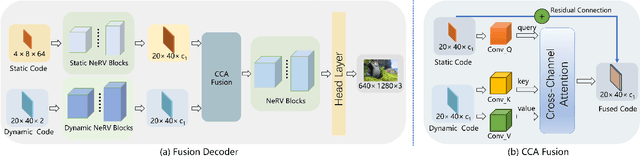
Abstract:Implicit neural representations for video (NeRV) have recently become a novel way for high-quality video representation. However, existing works employ a single network to represent the entire video, which implicitly confuse static and dynamic information. This leads to an inability to effectively compress the redundant static information and lack the explicitly modeling of global temporal-coherent dynamic details. To solve above problems, we propose DS-NeRV, which decomposes videos into sparse learnable static codes and dynamic codes without the need for explicit optical flow or residual supervision. By setting different sampling rates for two codes and applying weighted sum and interpolation sampling methods, DS-NeRV efficiently utilizes redundant static information while maintaining high-frequency details. Additionally, we design a cross-channel attention-based (CCA) fusion module to efficiently fuse these two codes for frame decoding. Our approach achieves a high quality reconstruction of 31.2 PSNR with only 0.35M parameters thanks to separate static and dynamic codes representation and outperforms existing NeRV methods in many downstream tasks. Our project website is at https://haoyan14.github.io/DS-NeRV.
Large language models in bioinformatics: applications and perspectives
Jan 08, 2024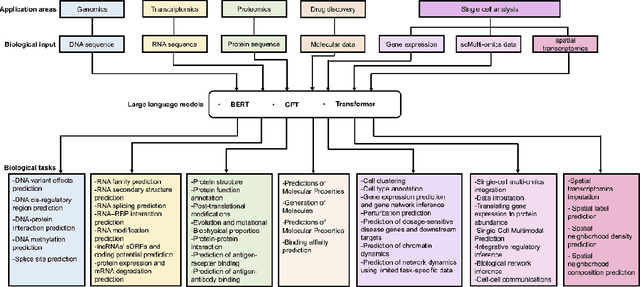
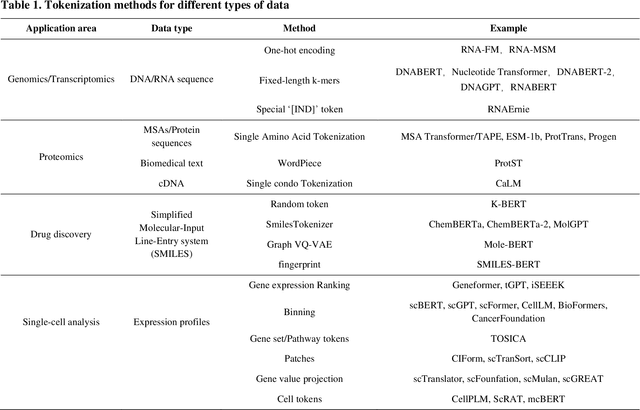
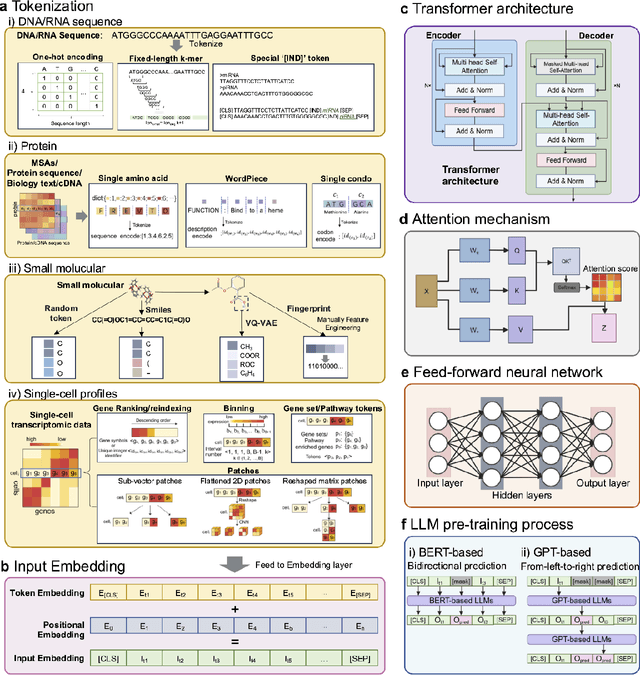
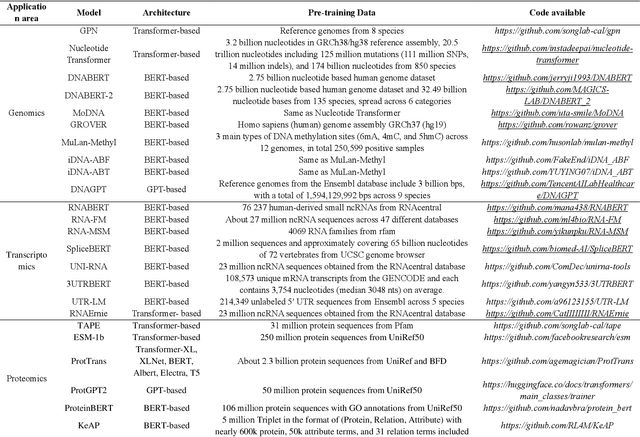
Abstract:Large language models (LLMs) are a class of artificial intelligence models based on deep learning, which have great performance in various tasks, especially in natural language processing (NLP). Large language models typically consist of artificial neural networks with numerous parameters, trained on large amounts of unlabeled input using self-supervised or semi-supervised learning. However, their potential for solving bioinformatics problems may even exceed their proficiency in modeling human language. In this review, we will present a summary of the prominent large language models used in natural language processing, such as BERT and GPT, and focus on exploring the applications of large language models at different omics levels in bioinformatics, mainly including applications of large language models in genomics, transcriptomics, proteomics, drug discovery and single cell analysis. Finally, this review summarizes the potential and prospects of large language models in solving bioinformatic problems.
4D-Editor: Interactive Object-level Editing in Dynamic Neural Radiance Fields via Semantic Distillation
Nov 06, 2023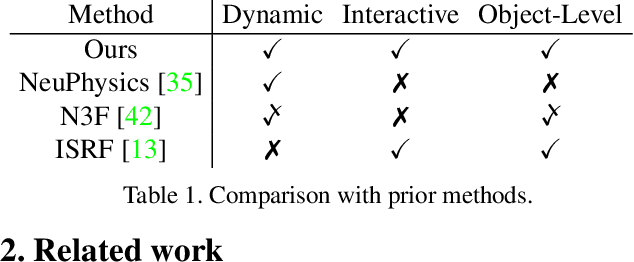
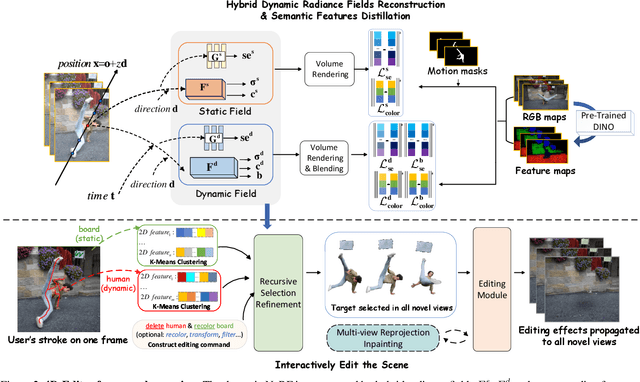
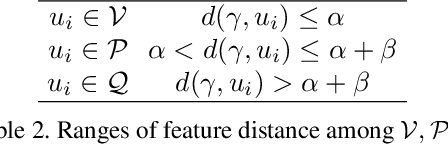
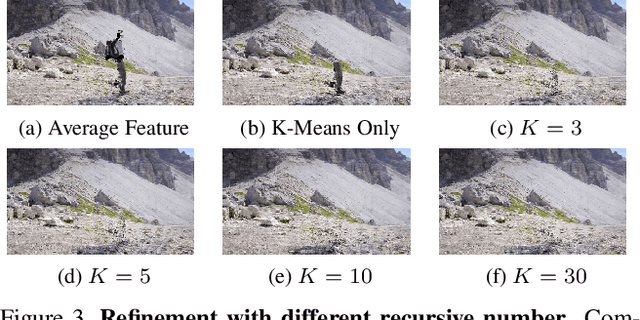
Abstract:This paper targets interactive object-level editing (e.g., deletion, recoloring, transformation, composition) in dynamic scenes. Recently, some methods aiming for flexible editing static scenes represented by neural radiance field (NeRF) have shown impressive synthesis quality, while similar capabilities in time-variant dynamic scenes remain limited. To solve this problem, we propose 4D-Editor, an interactive semantic-driven editing framework, allowing editing multiple objects in a dynamic NeRF with user strokes on a single frame. We propose an extension to the original dynamic NeRF by incorporating a hybrid semantic feature distillation to maintain spatial-temporal consistency after editing. In addition, we design Recursive Selection Refinement that significantly boosts object segmentation accuracy within a dynamic NeRF to aid the editing process. Moreover, we develop Multi-view Reprojection Inpainting to fill holes caused by incomplete scene capture after editing. Extensive experiments and editing examples on real-world demonstrate that 4D-Editor achieves photo-realistic editing on dynamic NeRFs. Project page: https://patrickddj.github.io/4D-Editor
AI assisted method for efficiently generating breast ultrasound screening reports
Jul 28, 2021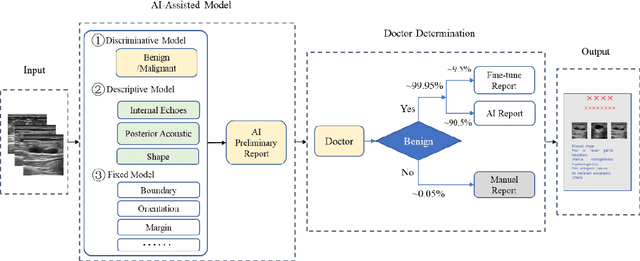

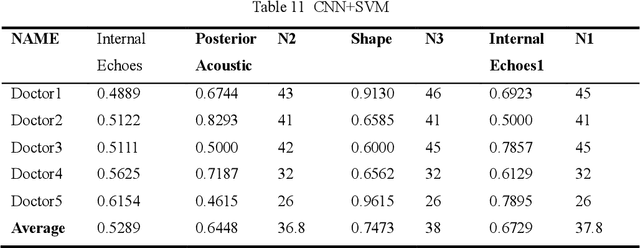
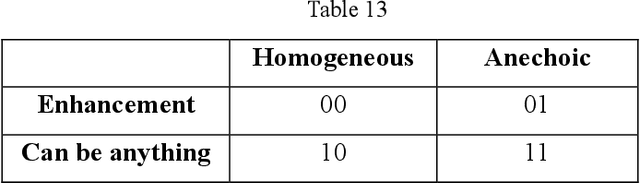
Abstract:Ultrasound is the preferred choice for early screening of dense breast cancer. Clinically, doctors have to manually write the screening report which is time-consuming and laborious, and it is easy to miss and miswrite. Therefore, this paper proposes a method for efficiently generating personalized breast ultrasound screening preliminary reports by AI, especially for benign and normal cases which account for the majority. Doctors then make simple adjustments or corrections to quickly generate final reports. The proposed approach has been tested using a database of 1133 breast tumor instances. Experimental results indicate this pipeline improves doctors' work efficiency by up to 90%, which greatly reduces repetitive work.
DC-Al GAN: Pseudoprogression and True Tumor Progression of Glioblastoma multiform Image Classification Based On DCGAN and Alexnet
Feb 26, 2019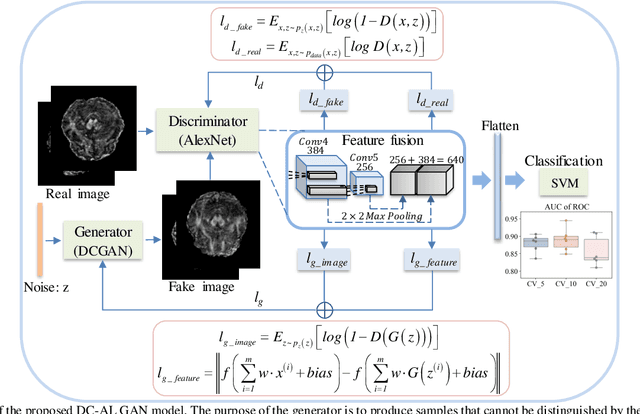
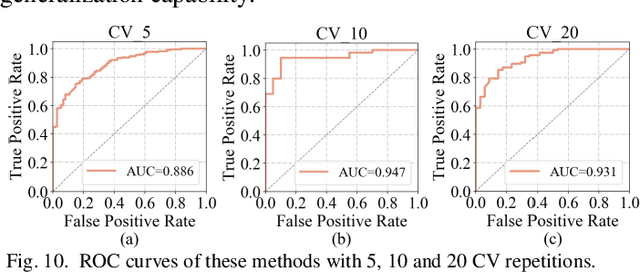
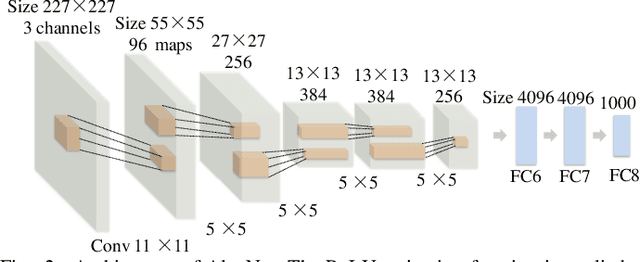
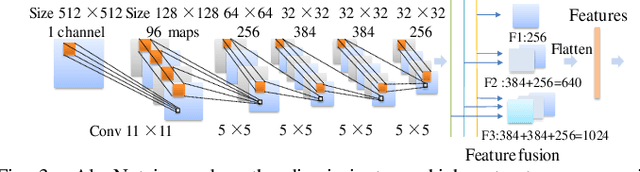
Abstract:Glioblastoma multiform (GBM) is a kind of head tumor with an extraordinarily complex treatment process. The survival period is typically 14-16 months, and the 2 year survival rate is approximately 26%-33%. The clinical treatment strategies for the pseudoprogression (PsP) and true tumor progression (TTP) of GBM are different, so accurately distinguishing these two conditions is particularly significant.As PsP and TTP of GBM are similar in shape and other characteristics, it is hard to distinguish these two forms with precision. In order to differentiate them accurately, this paper introduces a feature learning method based on a generative adversarial network: DC-Al GAN. GAN consists of two architectures: generator and discriminator. Alexnet is used as the discriminator in this work. Owing to the adversarial and competitive relationship between generator and discriminator, the latter extracts highly concise features during training. In DC-Al GAN, features are extracted from Alexnet in the final classification phase, and the highly nature of them contributes positively to the classification accuracy.The generator in DC-Al GAN is modified by the deep convolutional generative adversarial network (DCGAN) by adding three convolutional layers. This effectively generates higher resolution sample images. Feature fusion is used to combine high layer features with low layer features, allowing for the creation and use of more precise features for classification. The experimental results confirm that DC-Al GAN achieves high accuracy on GBM datasets for PsP and TTP image classification, which is superior to other state-of-the-art methods.
Efforts estimation of doctors annotating medical image
Jan 06, 2019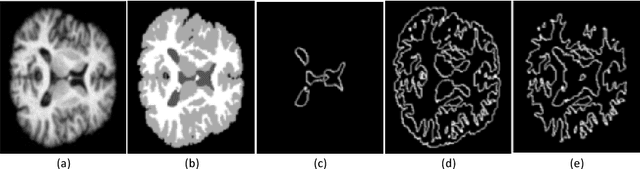
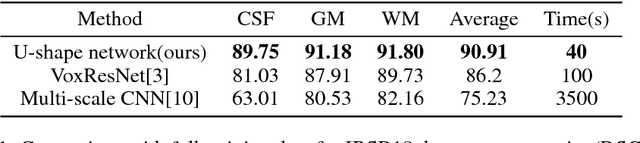
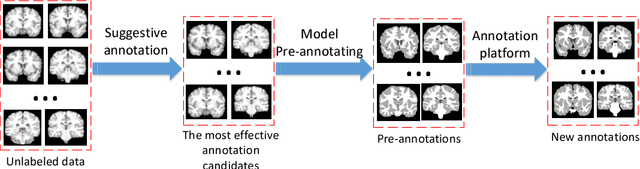
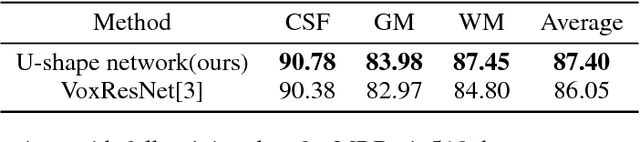
Abstract:Accurate annotation of medical image is the crucial step for image AI clinical application. However, annotating medical image will incur a great deal of annotation effort and expense due to its high complexity and needing experienced doctors. To alleviate annotation cost, some active learning methods are proposed. But such methods just cut the number of annotation candidates and do not study how many efforts the doctor will exactly take, which is not enough since even annotating a small amount of medical data will take a lot of time for the doctor. In this paper, we propose a new criterion to evaluate efforts of doctors annotating medical image. First, by coming active learning and U-shape network, we employ a suggestive annotation strategy to choose the most effective annotation candidates. Then we exploit a fine annotation platform to alleviate annotating efforts on each candidate and first utilize a new criterion to quantitatively calculate the efforts taken by doctors. In our work, we take MR brain tissue segmentation as an example to evaluate the proposed method. Extensive experiments on the well-known IBSR18 dataset and MRBrainS18 Challenge dataset show that, using proposed strategy, state-of-the-art segmentation performance can be achieved by using only 60% annotation candidates and annotation efforts can be alleviated by at least 44%, 44%, 47% on CSF, GM, WM separately.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge