Shuang Ge
Deep Learning in Single-Cell and Spatial Transcriptomics Data Analysis: Advances and Challenges from a Data Science Perspective
Dec 04, 2024



Abstract:The development of single-cell and spatial transcriptomics has revolutionized our capacity to investigate cellular properties, functions, and interactions in both cellular and spatial contexts. However, the analysis of single-cell and spatial omics data remains challenging. First, single-cell sequencing data are high-dimensional and sparse, often contaminated by noise and uncertainty, obscuring the underlying biological signals. Second, these data often encompass multiple modalities, including gene expression, epigenetic modifications, and spatial locations. Integrating these diverse data modalities is crucial for enhancing prediction accuracy and biological interpretability. Third, while the scale of single-cell sequencing has expanded to millions of cells, high-quality annotated datasets are still limited. Fourth, the complex correlations of biological tissues make it difficult to accurately reconstruct cellular states and spatial contexts. Traditional feature engineering-based analysis methods struggle to deal with the various challenges presented by intricate biological networks. Deep learning has emerged as a powerful tool capable of handling high-dimensional complex data and automatically identifying meaningful patterns, offering significant promise in addressing these challenges. This review systematically analyzes these challenges and discusses related deep learning approaches. Moreover, we have curated 21 datasets from 9 benchmarks, encompassing 58 computational methods, and evaluated their performance on the respective modeling tasks. Finally, we highlight three areas for future development from a technical, dataset, and application perspective. This work will serve as a valuable resource for understanding how deep learning can be effectively utilized in single-cell and spatial transcriptomics analyses, while inspiring novel approaches to address emerging challenges.
BSM loss: A superior way in modeling aleatory uncertainty of fine_grained classification
Jun 09, 2022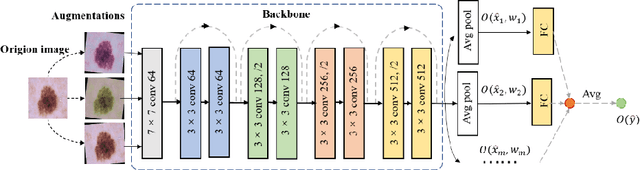
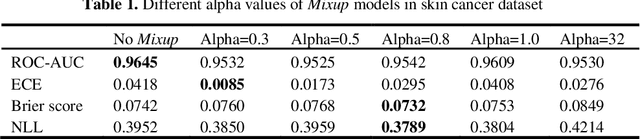
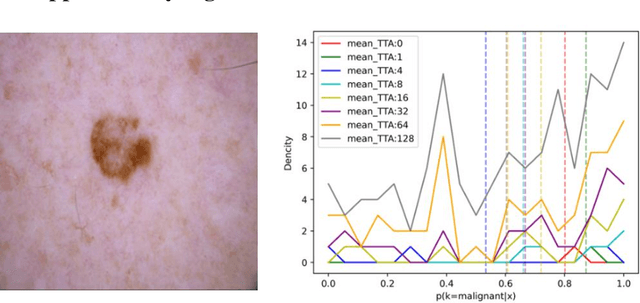
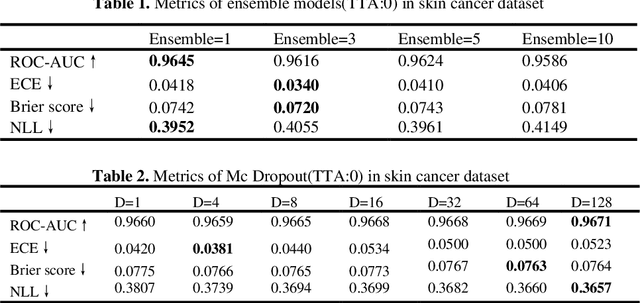
Abstract:Artificial intelligence(AI)-assisted method had received much attention in the risk field such as disease diagnosis. Different from the classification of disease types, it is a fine-grained task to classify the medical images as benign or malignant. However, most research only focuses on improving the diagnostic accuracy and ignores the evaluation of model reliability, which limits its clinical application. For clinical practice, calibration presents major challenges in the low-data regime extremely for over-parametrized models and inherent noises. In particular, we discovered that modeling data-dependent uncertainty is more conducive to confidence calibrations. Compared with test-time augmentation(TTA), we proposed a modified Bootstrapping loss(BS loss) function with Mixup data augmentation strategy that can better calibrate predictive uncertainty and capture data distribution transformation without additional inference time. Our experiments indicated that BS loss with Mixup(BSM) model can halve the Expected Calibration Error(ECE) compared to standard data augmentation, deep ensemble and MC dropout. The correlation between uncertainty and similarity of in-domain data is up to -0.4428 under the BSM model. Additionally, the BSM model is able to perceive the semantic distance of out-of-domain data, demonstrating high potential in real-world clinical practice.
MIPR:Automatic Annotation of Medical Images with Pixel Rearrangement
Apr 22, 2022



Abstract:Most of the state-of-the-art semantic segmentation reported in recent years is based on fully supervised deep learning in the medical domain. How?ever, the high-quality annotated datasets require intense labor and domain knowledge, consuming enormous time and cost. Previous works that adopt semi?supervised and unsupervised learning are proposed to address the lack of anno?tated data through assisted training with unlabeled data and achieve good perfor?mance. Still, these methods can not directly get the image annotation as doctors do. In this paper, inspired by self-training of semi-supervised learning, we pro?pose a novel approach to solve the lack of annotated data from another angle, called medical image pixel rearrangement (short in MIPR). The MIPR combines image-editing and pseudo-label technology to obtain labeled data. As the number of iterations increases, the edited image is similar to the original image, and the labeled result is similar to the doctor annotation. Therefore, the MIPR is to get labeled pairs of data directly from amounts of unlabled data with pixel rearrange?ment, which is implemented with a designed conditional Generative Adversarial Networks and a segmentation network. Experiments on the ISIC18 show that the effect of the data annotated by our method for segmentation task is is equal to or even better than that of doctors annotations
AI assisted method for efficiently generating breast ultrasound screening reports
Jul 28, 2021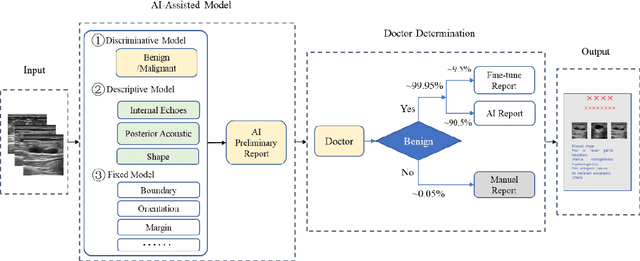

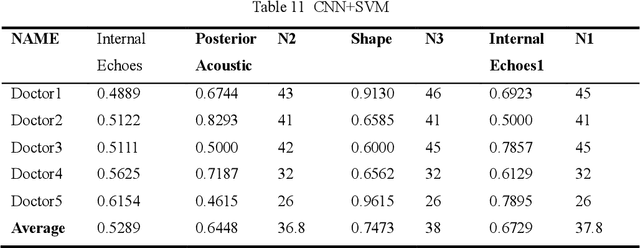
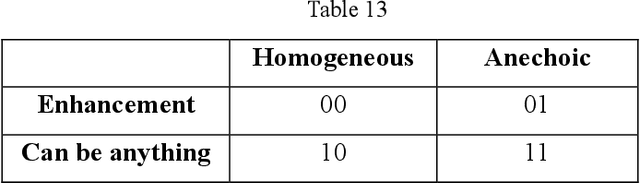
Abstract:Ultrasound is the preferred choice for early screening of dense breast cancer. Clinically, doctors have to manually write the screening report which is time-consuming and laborious, and it is easy to miss and miswrite. Therefore, this paper proposes a method for efficiently generating personalized breast ultrasound screening preliminary reports by AI, especially for benign and normal cases which account for the majority. Doctors then make simple adjustments or corrections to quickly generate final reports. The proposed approach has been tested using a database of 1133 breast tumor instances. Experimental results indicate this pipeline improves doctors' work efficiency by up to 90%, which greatly reduces repetitive work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge