Mengyuan Yang
TermGPT: Multi-Level Contrastive Fine-Tuning for Terminology Adaptation in Legal and Financial Domain
Nov 13, 2025Abstract:Large language models (LLMs) have demonstrated impressive performance in text generation tasks; however, their embedding spaces often suffer from the isotropy problem, resulting in poor discrimination of domain-specific terminology, particularly in legal and financial contexts. This weakness in terminology-level representation can severely hinder downstream tasks such as legal judgment prediction or financial risk analysis, where subtle semantic distinctions are critical. To address this problem, we propose TermGPT, a multi-level contrastive fine-tuning framework designed for terminology adaptation. We first construct a sentence graph to capture semantic and structural relations, and generate semantically consistent yet discriminative positive and negative samples based on contextual and topological cues. We then devise a multi-level contrastive learning approach at both the sentence and token levels, enhancing global contextual understanding and fine-grained terminology discrimination. To support robust evaluation, we construct the first financial terminology dataset derived from official regulatory documents. Experiments show that TermGPT outperforms existing baselines in term discrimination tasks within the finance and legal domains.
SPAST: Arbitrary Style Transfer with Style Priors via Pre-trained Large-scale Model
May 13, 2025Abstract:Given an arbitrary content and style image, arbitrary style transfer aims to render a new stylized image which preserves the content image's structure and possesses the style image's style. Existing arbitrary style transfer methods are based on either small models or pre-trained large-scale models. The small model-based methods fail to generate high-quality stylized images, bringing artifacts and disharmonious patterns. The pre-trained large-scale model-based methods can generate high-quality stylized images but struggle to preserve the content structure and cost long inference time. To this end, we propose a new framework, called SPAST, to generate high-quality stylized images with less inference time. Specifically, we design a novel Local-global Window Size Stylization Module (LGWSSM)tofuse style features into content features. Besides, we introduce a novel style prior loss, which can dig out the style priors from a pre-trained large-scale model into the SPAST and motivate the SPAST to generate high-quality stylized images with short inference time.We conduct abundant experiments to verify that our proposed method can generate high-quality stylized images and less inference time compared with the SOTA arbitrary style transfer methods.
DyArtbank: Diverse Artistic Style Transfer via Pre-trained Stable Diffusion and Dynamic Style Prompt Artbank
Mar 11, 2025



Abstract:Artistic style transfer aims to transfer the learned style onto an arbitrary content image. However, most existing style transfer methods can only render consistent artistic stylized images, making it difficult for users to get enough stylized images to enjoy. To solve this issue, we propose a novel artistic style transfer framework called DyArtbank, which can generate diverse and highly realistic artistic stylized images. Specifically, we introduce a Dynamic Style Prompt ArtBank (DSPA), a set of learnable parameters. It can learn and store the style information from the collection of artworks, dynamically guiding pre-trained stable diffusion to generate diverse and highly realistic artistic stylized images. DSPA can also generate random artistic image samples with the learned style information, providing a new idea for data augmentation. Besides, a Key Content Feature Prompt (KCFP) module is proposed to provide sufficient content prompts for pre-trained stable diffusion to preserve the detailed structure of the input content image. Extensive qualitative and quantitative experiments verify the effectiveness of our proposed method. Code is available: https://github.com/Jamie-Cheung/DyArtbank
Large language models in bioinformatics: applications and perspectives
Jan 08, 2024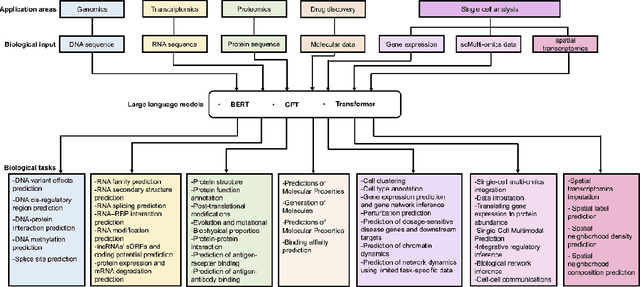
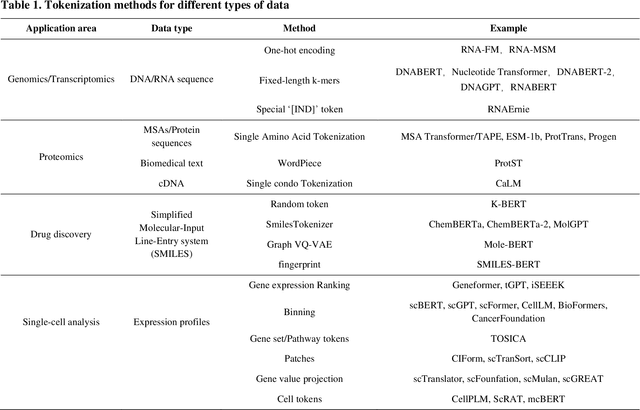
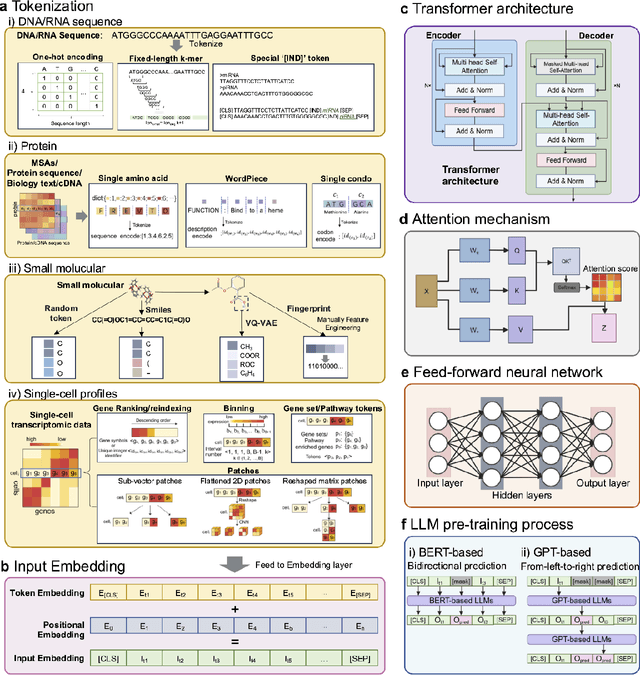
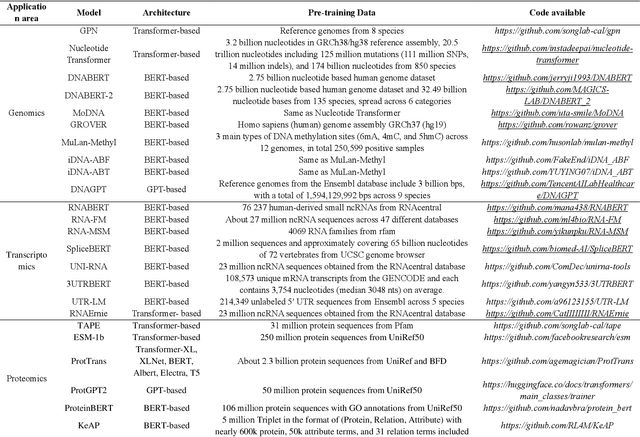
Abstract:Large language models (LLMs) are a class of artificial intelligence models based on deep learning, which have great performance in various tasks, especially in natural language processing (NLP). Large language models typically consist of artificial neural networks with numerous parameters, trained on large amounts of unlabeled input using self-supervised or semi-supervised learning. However, their potential for solving bioinformatics problems may even exceed their proficiency in modeling human language. In this review, we will present a summary of the prominent large language models used in natural language processing, such as BERT and GPT, and focus on exploring the applications of large language models at different omics levels in bioinformatics, mainly including applications of large language models in genomics, transcriptomics, proteomics, drug discovery and single cell analysis. Finally, this review summarizes the potential and prospects of large language models in solving bioinformatic problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge