Mingwang Zhu
Efforts estimation of doctors annotating medical image
Jan 06, 2019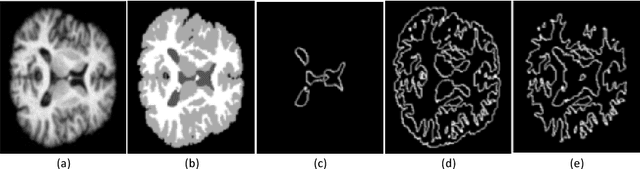
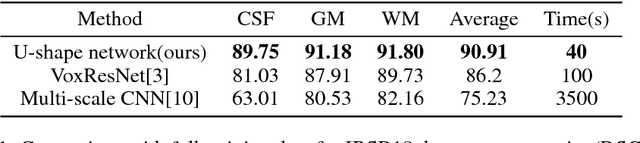
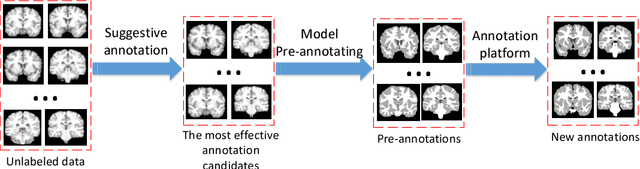
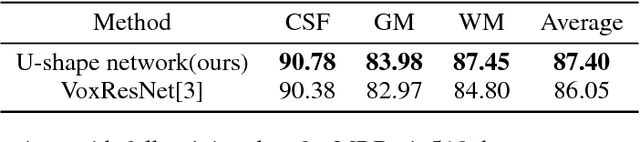
Abstract:Accurate annotation of medical image is the crucial step for image AI clinical application. However, annotating medical image will incur a great deal of annotation effort and expense due to its high complexity and needing experienced doctors. To alleviate annotation cost, some active learning methods are proposed. But such methods just cut the number of annotation candidates and do not study how many efforts the doctor will exactly take, which is not enough since even annotating a small amount of medical data will take a lot of time for the doctor. In this paper, we propose a new criterion to evaluate efforts of doctors annotating medical image. First, by coming active learning and U-shape network, we employ a suggestive annotation strategy to choose the most effective annotation candidates. Then we exploit a fine annotation platform to alleviate annotating efforts on each candidate and first utilize a new criterion to quantitatively calculate the efforts taken by doctors. In our work, we take MR brain tissue segmentation as an example to evaluate the proposed method. Extensive experiments on the well-known IBSR18 dataset and MRBrainS18 Challenge dataset show that, using proposed strategy, state-of-the-art segmentation performance can be achieved by using only 60% annotation candidates and annotation efforts can be alleviated by at least 44%, 44%, 47% on CSF, GM, WM separately.
A Multi-channel Network with Image Retrieval for Accurate Brain Tissue Segmentation
Aug 04, 2018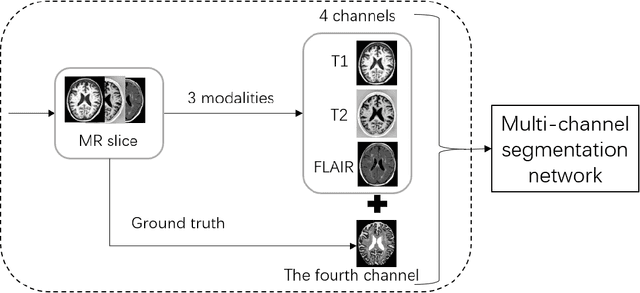

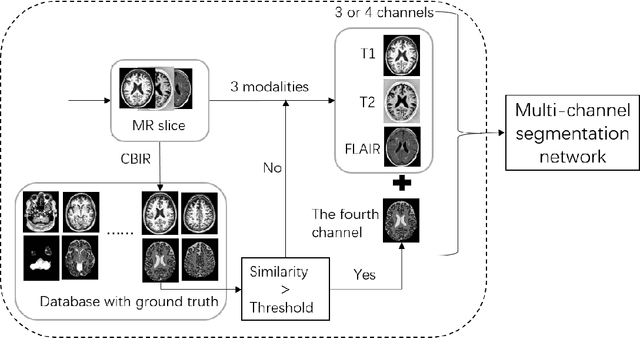
Abstract:Magnetic Resonance Imaging (MRI) is widely used in the pathological and functional studies of the brain, such as epilepsy, tumor diagnosis, etc. Automated accurate brain tissue segmentation like cerebro-spinal fluid (CSF), gray matter (GM), white matter (WM) is the basis of these studies and many researchers are seeking it to the best. Based on the truth that multi-channel segmentation network with its own ground truth achieves up to average dice ratio 0.98, we propose a novel method that we add a fourth channel with the ground truth of the most similar image's obtained by CBIR from the database. The results show that the method improves the segmentation performance, as measured by average dice ratio, by approximately 0.01 in the MRBrainS18 database. In addition, our method is concise and robust, which can be used to any network architecture that needs not be modified a lot.
A Strategy of MR Brain Tissue Images' Suggestive Annotation Based on Modified U-Net
Jul 28, 2018
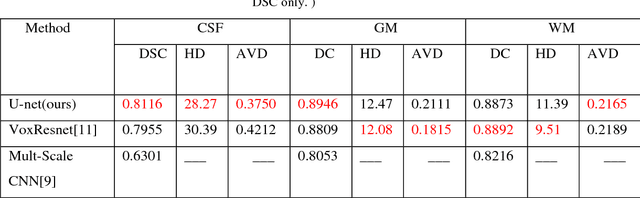
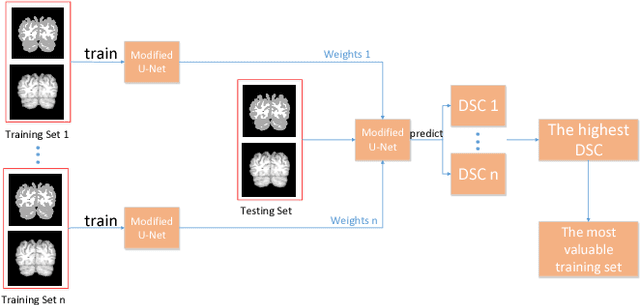
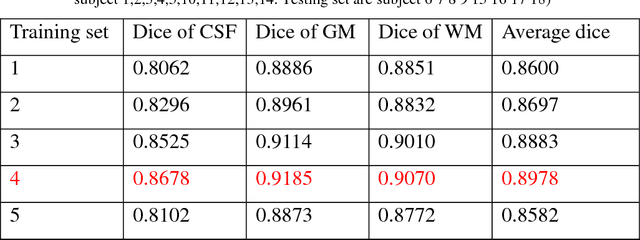
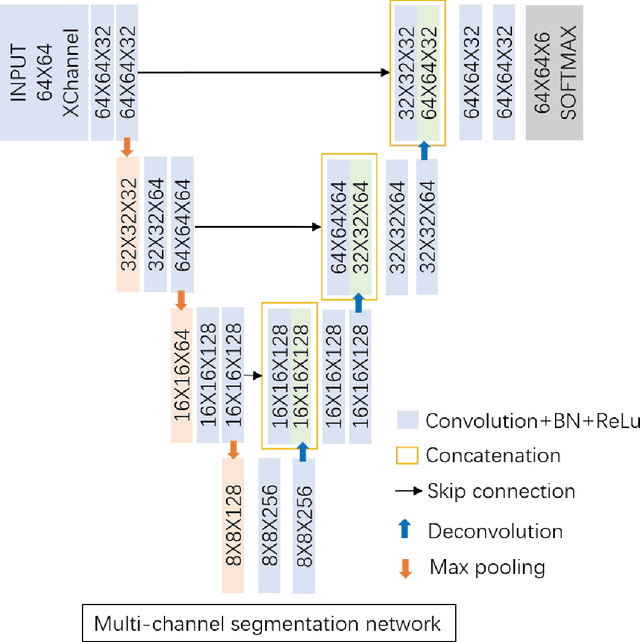
Abstract:Accurate segmentation of MR brain tissue is a crucial step for diagnosis,surgical planning, and treatment of brain abnormalities. However,it is a time-consuming task to be performed by medical experts. So, automatic and reliable segmentation methods are required. How to choose appropriate training dataset from limited labeled dataset rather than the whole also has great significance in saving training time. In addition, medical data labeled is too rare and expensive to obtain extensively, so choosing appropriate unlabeled dataset instead of all the datasets to annotate, which can attain at least same performance, is also very meaningful. To solve the problem above, we design an automatic segmentation method based on U-shaped deep convolutional network and obtain excellent result with average DSC metric of 0.8610, 0.9131, 0.9003 for Cerebrospinal Fluid (CSF), Gray Matter (GM) and White Matter (WM) respectively on the well-known IBSR18 dataset. We use bootstrapping algorithm for selecting the most effective training data and get more state-of-the-art segmentation performance by using only 50% of training data. Moreover, we propose a strategy of MR brain tissue images' suggestive annotation for unlabeled medical data based on the modified U-net. The proposed method performs fast and can be used in clinical.
DASN:Data-Aware Skilled Network for Accurate MR Brain Tissue Segmentation
Jul 24, 2018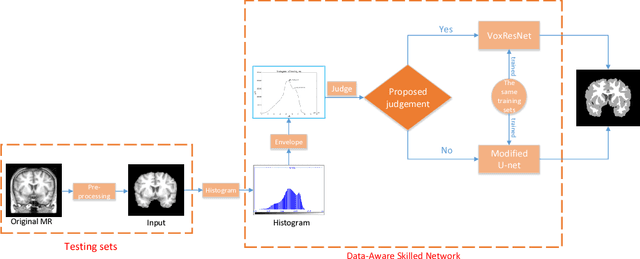
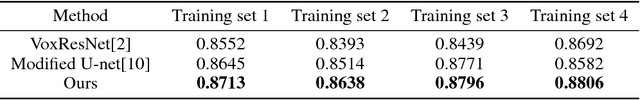
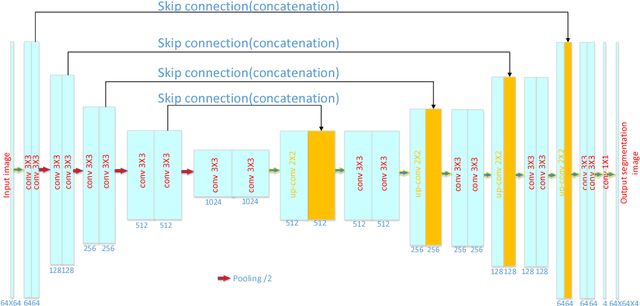
Abstract:Accurate segmentation of MR brain tissue is a crucial step for diagnosis, surgical planning, and treatment of brain abnormalities. Automatic and reliable segmenta-tion methods are required to assist doctor. Over the last few years, deep learning especially deep convolutional neural networks (CNNs) have emerged as one of the most prominent approaches for image recognition problems in various do-mains. But the improvement of deep networks always needs inspiration, which is rare for the ordinary. Until now,there have been reasonable MR brain tissue segmentation methods,all of which can achieve promising performance. These different methods have their own characteristic and are distinctive for data sets. In other words, different models performance vary widely on the same data sets and each model has what it is skilled in. It is on the basis of this, we propose a judgement to distinguish data sets that different models are good at. With our method, the segmentation accuracy can be improved easily based on the existing models, neither without increasing training data nor improving the network. We validate our method on the widely used IBSR 18 dataset and obtain average dice ratio of 88.06%,while it is 85.82% and 86.92% when only using separate one model respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge