Yongpei Zhu
Interpretable ODE-style Generative Diffusion Model via Force Field Construction
Mar 15, 2023Abstract:For a considerable time, researchers have focused on developing a method that establishes a deep connection between the generative diffusion model and mathematical physics. Despite previous efforts, progress has been limited to the pursuit of a single specialized method. In order to advance the interpretability of diffusion models and explore new research directions, it is essential to establish a unified ODE-style generative diffusion model. Such a model should draw inspiration from physical models and possess a clear geometric meaning. This paper aims to identify various physical models that are suitable for constructing ODE-style generative diffusion models accurately from a mathematical perspective. We then summarize these models into a unified method. Additionally, we perform a case study where we use the theoretical model identified by our method to develop a range of new diffusion model methods, and conduct experiments. Our experiments on CIFAR-10 demonstrate the effectiveness of our approach. We have constructed a computational framework that attains highly proficient results with regards to image generation speed, alongside an additional model that demonstrates exceptional performance in both Inception score and FID score. These results underscore the significance of our method in advancing the field of diffusion models.
Deformable Registration Using Average Geometric Transformations for Brain MR Images
Jul 23, 2019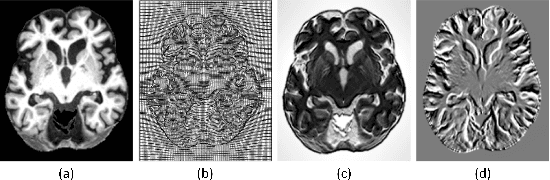
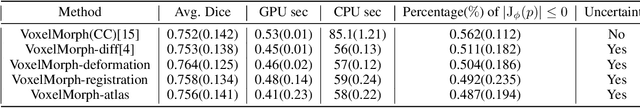
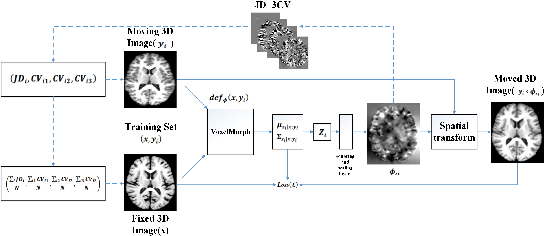
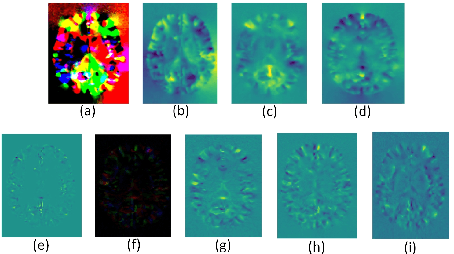
Abstract:Accurate registration of medical images is vital for doctor's diagnosis and quantitative analysis. In this paper, we propose a new deformable medical image registration method based on average geometric transformations and VoxelMorph CNN architecture. We compute the differential geometric information including Jacobian determinant(JD) and the curl vector(CV) of diffeomorphic registration field and use them as multi-channel of VoxelMorph CNN for second train. In addition, we use the average transformation to construct a standard brain MRI atlas which can be used as fixed image. We verify our method on two datasets including ADNI dataset and MRBrainS18 Challenge dataset, and obtain excellent improvement on MR image registration with average Dice scores and non-negative Jacobian locations compared with MIT's original method. The experimental results show the method can achieve better performance in brain MRI diagnosis.
Facial Expression Recognition Research Based on Deep Learning
Apr 24, 2019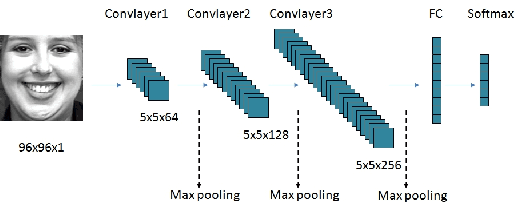
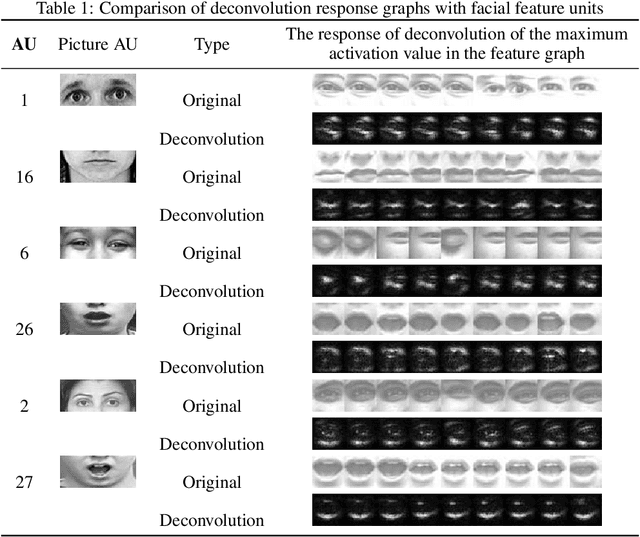

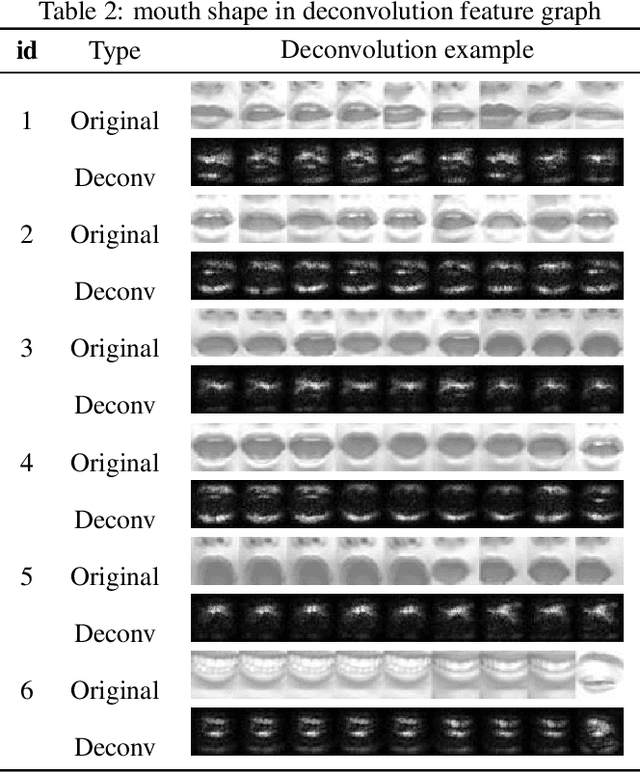
Abstract:With the development of deep learning, the structure of convolution neural network is becoming more and more complex and the performance of object recognition is getting better. However, the classification mechanism of convolution neural networks is still an unsolved core problem. The main problem is that convolution neural networks have too many parameters, which makes it difficult to analyze them. In this paper, we design and train a convolution neural network based on the expression recognition, and explore the classification mechanism of the network. By using the Deconvolution visualization method, the extremum point of the convolution neural network is projected back to the pixel space of the original image, and we qualitatively verify that the trained expression recognition convolution neural network forms a detector for the specific facial action unit. At the same time, we design the distance function to measure the distance between the presence of facial feature unit and the maximal value of the response on the feature map of convolution neural network. The greater the distance, the more sensitive the feature map is to the facial feature unit. By comparing the maximum distance of all facial feature elements in the feature graph, the mapping relationship between facial feature element and convolution neural network feature map is determined. Therefore, we have verified that the convolution neural network has formed a detector for the facial Action unit in the training process to realize the expression recognition.
The Generation and Application of Medical Image Grid Based on Differential Geometric Features
Jan 30, 2019

Abstract:Accurate segmentation of brain tissue in magnetic resonance images (MRI) is a difficult task due to different types of brain abnormalities. In this paper, we review the deformation method focus on the construction of diffeomorphisms, address clearly a new formation of the deformation problem for moving domains, and we apply it in natural images, face images and MRI brain images. And we use a new method to construct diffeomorphisms through a completely different approach. The idea is to control directly the Jacobian determinant and the curl vector of a transformation and use them as one CNN channel with other modalities(T1-weighted, T1-IR and T2-FLAIR) to get more accurate results of brain segmentation. More importantly, we discuss the influence of some optimization parameters to precision analysis of MRI brain segmentation by both numerical experiments and theoretical analysis. We test this method on the IBSR dataset and MRBrainS18 dataset based on VoxResNet and prove the influence of three parameters on the accuracy of MRI brain segmentation.Finally, we also compare the segmentation performance of our method in two networks, VoxResNet and 3D U-Net network. We believe the proposed method can advance the performance in brain segmentation and clinical diagnosis.
Efforts estimation of doctors annotating medical image
Jan 06, 2019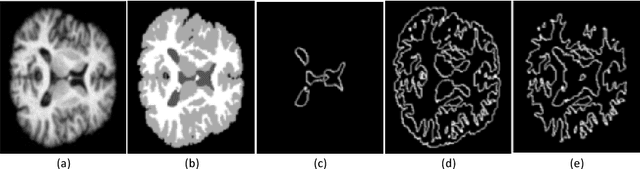
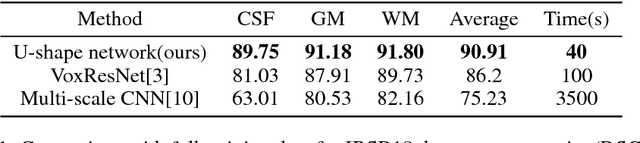
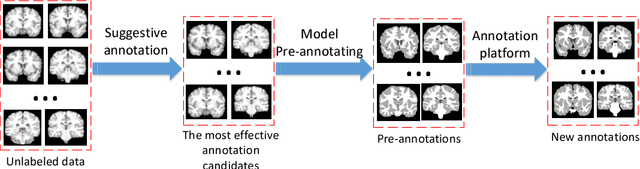
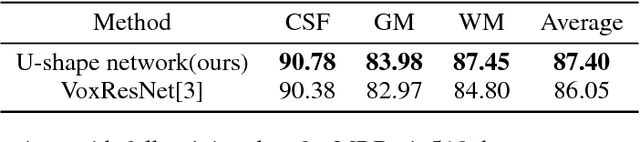
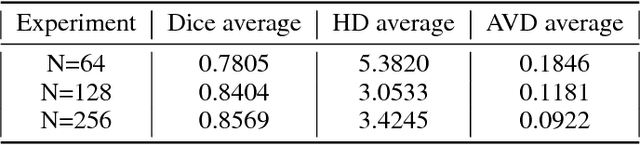
Abstract:Accurate annotation of medical image is the crucial step for image AI clinical application. However, annotating medical image will incur a great deal of annotation effort and expense due to its high complexity and needing experienced doctors. To alleviate annotation cost, some active learning methods are proposed. But such methods just cut the number of annotation candidates and do not study how many efforts the doctor will exactly take, which is not enough since even annotating a small amount of medical data will take a lot of time for the doctor. In this paper, we propose a new criterion to evaluate efforts of doctors annotating medical image. First, by coming active learning and U-shape network, we employ a suggestive annotation strategy to choose the most effective annotation candidates. Then we exploit a fine annotation platform to alleviate annotating efforts on each candidate and first utilize a new criterion to quantitatively calculate the efforts taken by doctors. In our work, we take MR brain tissue segmentation as an example to evaluate the proposed method. Extensive experiments on the well-known IBSR18 dataset and MRBrainS18 Challenge dataset show that, using proposed strategy, state-of-the-art segmentation performance can be achieved by using only 60% annotation candidates and annotation efforts can be alleviated by at least 44%, 44%, 47% on CSF, GM, WM separately.
The Method of Multimodal MRI Brain Image Segmentation Based on Differential Geometric Features
Nov 28, 2018
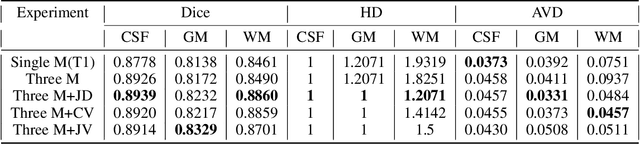
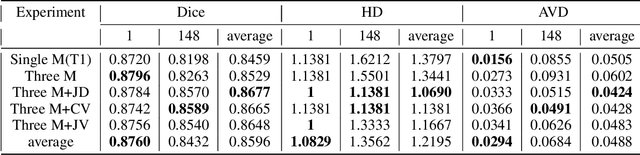
Abstract:Accurate segmentation of brain tissue in magnetic resonance images (MRI) is a diffcult task due to different types of brain abnormalities. Using information and features from multimodal MRI including T1, T1-weighted inversion recovery (T1-IR) and T2-FLAIR and differential geometric features including the Jacobian determinant(JD) and the curl vector(CV) derived from T1 modality can result in a more accurate analysis of brain images. In this paper, we use the differential geometric information including JD and CV as image characteristics to measure the differences between different MRI images, which represent local size changes and local rotations of the brain image, and we can use them as one CNN channel with other three modalities (T1-weighted, T1-IR and T2-FLAIR) to get more accurate results of brain segmentation. We test this method on two datasets including IBSR dataset and MRBrainS datasets based on the deep voxelwise residual network, namely VoxResNet, and obtain excellent improvement over single modality or three modalities and increases average DSC(Cerebrospinal Fluid (CSF), Gray Matter (GM) and White Matter (WM)) by about 1.5% on the well-known MRBrainS18 dataset and about 2.5% on the IBSR dataset. Moreover, we discuss that one modality combined with its JD or CV information can replace the segmentation effect of three modalities, which can provide medical conveniences for doctor to diagnose because only to extract T1-modality MRI image of patients. Finally, we also compare the segmentation performance of our method in two networks, VoxResNet and U-Net network. The results show VoxResNet has a better performance than U-Net network with our method in brain MRI segmentation. We believe the proposed method can advance the performance in brain segmentation and clinical diagnosis.
A Strategy of MR Brain Tissue Images' Suggestive Annotation Based on Modified U-Net
Jul 28, 2018
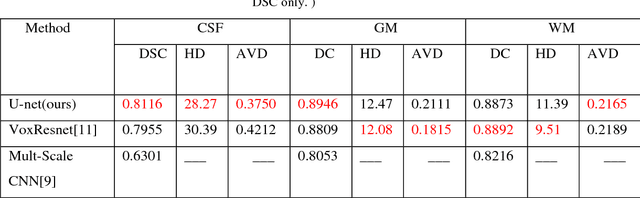
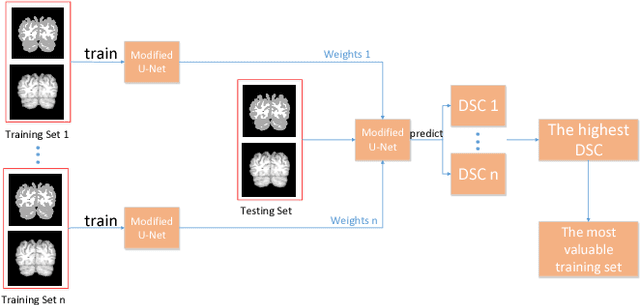
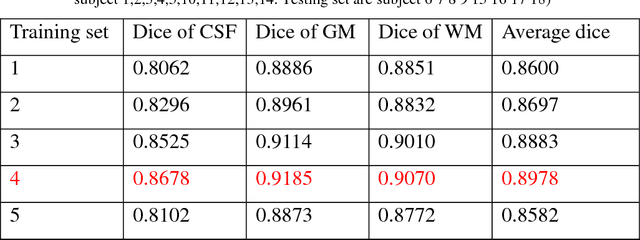
Abstract:Accurate segmentation of MR brain tissue is a crucial step for diagnosis,surgical planning, and treatment of brain abnormalities. However,it is a time-consuming task to be performed by medical experts. So, automatic and reliable segmentation methods are required. How to choose appropriate training dataset from limited labeled dataset rather than the whole also has great significance in saving training time. In addition, medical data labeled is too rare and expensive to obtain extensively, so choosing appropriate unlabeled dataset instead of all the datasets to annotate, which can attain at least same performance, is also very meaningful. To solve the problem above, we design an automatic segmentation method based on U-shaped deep convolutional network and obtain excellent result with average DSC metric of 0.8610, 0.9131, 0.9003 for Cerebrospinal Fluid (CSF), Gray Matter (GM) and White Matter (WM) respectively on the well-known IBSR18 dataset. We use bootstrapping algorithm for selecting the most effective training data and get more state-of-the-art segmentation performance by using only 50% of training data. Moreover, we propose a strategy of MR brain tissue images' suggestive annotation for unlabeled medical data based on the modified U-net. The proposed method performs fast and can be used in clinical.
DASN:Data-Aware Skilled Network for Accurate MR Brain Tissue Segmentation
Jul 24, 2018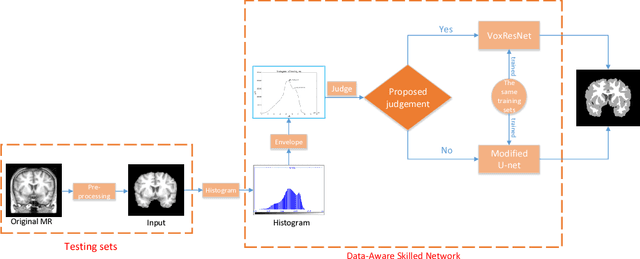
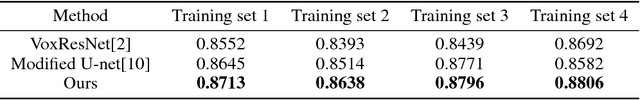
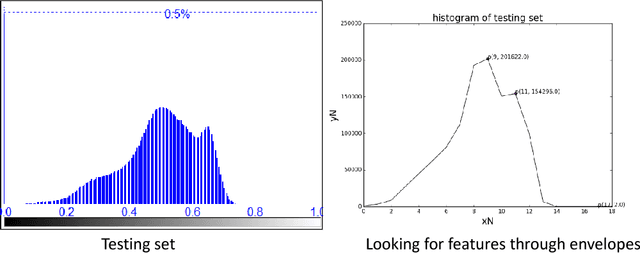
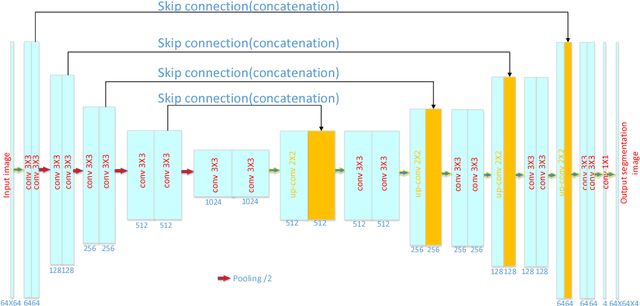
Abstract:Accurate segmentation of MR brain tissue is a crucial step for diagnosis, surgical planning, and treatment of brain abnormalities. Automatic and reliable segmenta-tion methods are required to assist doctor. Over the last few years, deep learning especially deep convolutional neural networks (CNNs) have emerged as one of the most prominent approaches for image recognition problems in various do-mains. But the improvement of deep networks always needs inspiration, which is rare for the ordinary. Until now,there have been reasonable MR brain tissue segmentation methods,all of which can achieve promising performance. These different methods have their own characteristic and are distinctive for data sets. In other words, different models performance vary widely on the same data sets and each model has what it is skilled in. It is on the basis of this, we propose a judgement to distinguish data sets that different models are good at. With our method, the segmentation accuracy can be improved easily based on the existing models, neither without increasing training data nor improving the network. We validate our method on the widely used IBSR 18 dataset and obtain average dice ratio of 88.06%,while it is 85.82% and 86.92% when only using separate one model respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge