Zicong Zhou
Deformable Registration Using Average Geometric Transformations for Brain MR Images
Jul 23, 2019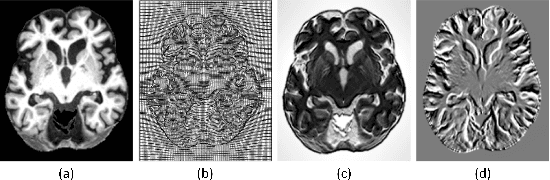
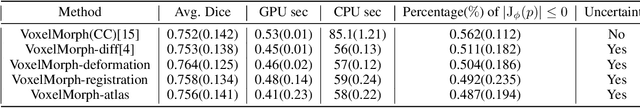
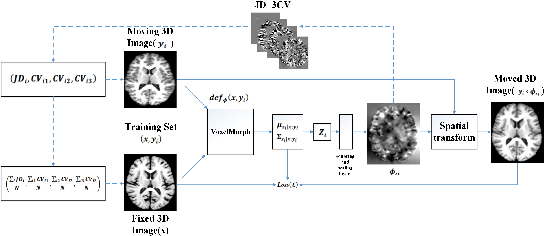
Abstract:Accurate registration of medical images is vital for doctor's diagnosis and quantitative analysis. In this paper, we propose a new deformable medical image registration method based on average geometric transformations and VoxelMorph CNN architecture. We compute the differential geometric information including Jacobian determinant(JD) and the curl vector(CV) of diffeomorphic registration field and use them as multi-channel of VoxelMorph CNN for second train. In addition, we use the average transformation to construct a standard brain MRI atlas which can be used as fixed image. We verify our method on two datasets including ADNI dataset and MRBrainS18 Challenge dataset, and obtain excellent improvement on MR image registration with average Dice scores and non-negative Jacobian locations compared with MIT's original method. The experimental results show the method can achieve better performance in brain MRI diagnosis.
The Generation and Application of Medical Image Grid Based on Differential Geometric Features
Jan 30, 2019

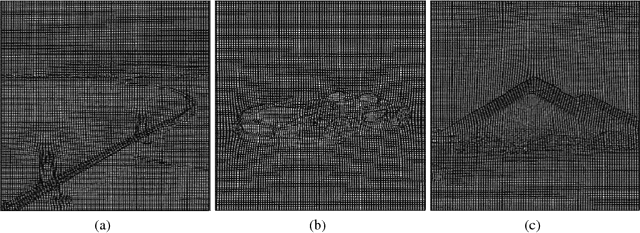
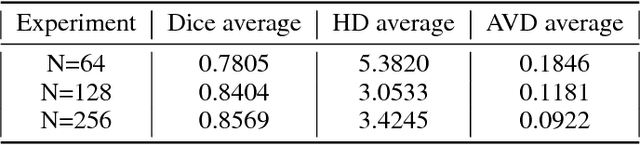
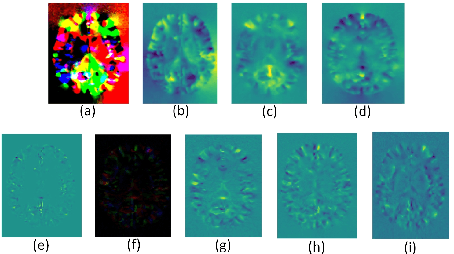
Abstract:Accurate segmentation of brain tissue in magnetic resonance images (MRI) is a difficult task due to different types of brain abnormalities. In this paper, we review the deformation method focus on the construction of diffeomorphisms, address clearly a new formation of the deformation problem for moving domains, and we apply it in natural images, face images and MRI brain images. And we use a new method to construct diffeomorphisms through a completely different approach. The idea is to control directly the Jacobian determinant and the curl vector of a transformation and use them as one CNN channel with other modalities(T1-weighted, T1-IR and T2-FLAIR) to get more accurate results of brain segmentation. More importantly, we discuss the influence of some optimization parameters to precision analysis of MRI brain segmentation by both numerical experiments and theoretical analysis. We test this method on the IBSR dataset and MRBrainS18 dataset based on VoxResNet and prove the influence of three parameters on the accuracy of MRI brain segmentation.Finally, we also compare the segmentation performance of our method in two networks, VoxResNet and 3D U-Net network. We believe the proposed method can advance the performance in brain segmentation and clinical diagnosis.
The Method of Multimodal MRI Brain Image Segmentation Based on Differential Geometric Features
Nov 28, 2018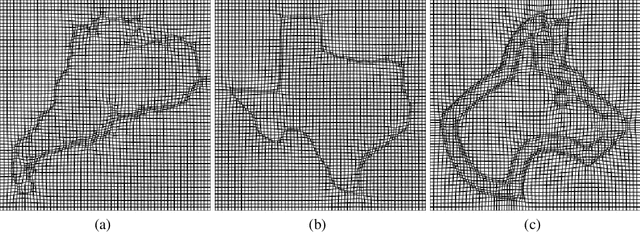
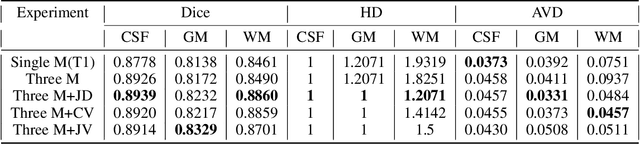
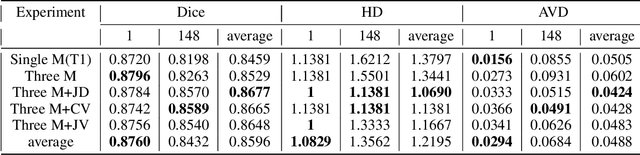
Abstract:Accurate segmentation of brain tissue in magnetic resonance images (MRI) is a diffcult task due to different types of brain abnormalities. Using information and features from multimodal MRI including T1, T1-weighted inversion recovery (T1-IR) and T2-FLAIR and differential geometric features including the Jacobian determinant(JD) and the curl vector(CV) derived from T1 modality can result in a more accurate analysis of brain images. In this paper, we use the differential geometric information including JD and CV as image characteristics to measure the differences between different MRI images, which represent local size changes and local rotations of the brain image, and we can use them as one CNN channel with other three modalities (T1-weighted, T1-IR and T2-FLAIR) to get more accurate results of brain segmentation. We test this method on two datasets including IBSR dataset and MRBrainS datasets based on the deep voxelwise residual network, namely VoxResNet, and obtain excellent improvement over single modality or three modalities and increases average DSC(Cerebrospinal Fluid (CSF), Gray Matter (GM) and White Matter (WM)) by about 1.5% on the well-known MRBrainS18 dataset and about 2.5% on the IBSR dataset. Moreover, we discuss that one modality combined with its JD or CV information can replace the segmentation effect of three modalities, which can provide medical conveniences for doctor to diagnose because only to extract T1-modality MRI image of patients. Finally, we also compare the segmentation performance of our method in two networks, VoxResNet and U-Net network. The results show VoxResNet has a better performance than U-Net network with our method in brain MRI segmentation. We believe the proposed method can advance the performance in brain segmentation and clinical diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge