Saima Rathore
A Recent Survey of Vision Transformers for Medical Image Segmentation
Dec 01, 2023
Abstract:Medical image segmentation plays a crucial role in various healthcare applications, enabling accurate diagnosis, treatment planning, and disease monitoring. In recent years, Vision Transformers (ViTs) have emerged as a promising technique for addressing the challenges in medical image segmentation. In medical images, structures are usually highly interconnected and globally distributed. ViTs utilize their multi-scale attention mechanism to model the long-range relationships in the images. However, they do lack image-related inductive bias and translational invariance, potentially impacting their performance. Recently, researchers have come up with various ViT-based approaches that incorporate CNNs in their architectures, known as Hybrid Vision Transformers (HVTs) to capture local correlation in addition to the global information in the images. This survey paper provides a detailed review of the recent advancements in ViTs and HVTs for medical image segmentation. Along with the categorization of ViT and HVT-based medical image segmentation approaches we also present a detailed overview of their real-time applications in several medical image modalities. This survey may serve as a valuable resource for researchers, healthcare practitioners, and students in understanding the state-of-the-art approaches for ViT-based medical image segmentation.
Deep radiomic signature with immune cell markers predicts the survival of glioma patients
Jun 09, 2022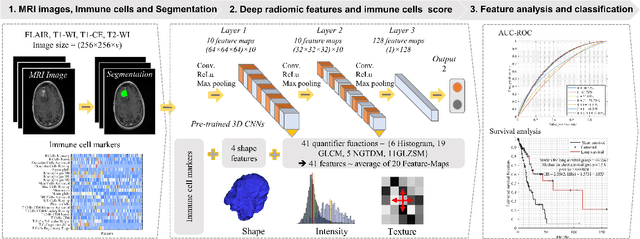
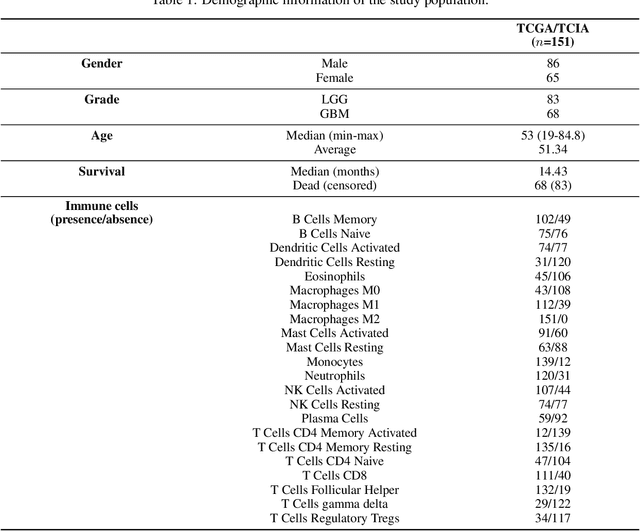
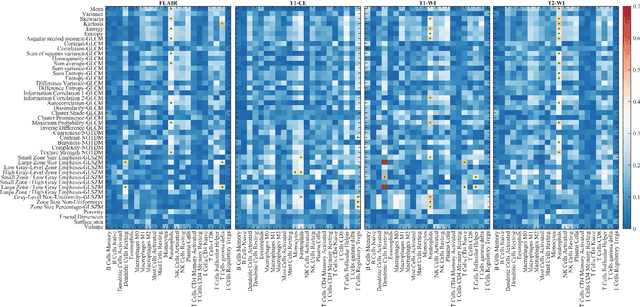
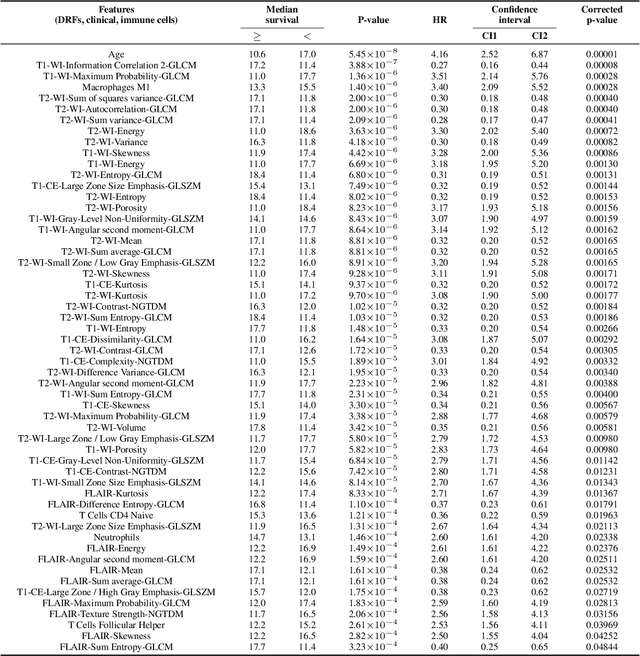
Abstract:Imaging biomarkers offer a non-invasive way to predict the response of immunotherapy prior to treatment. In this work, we propose a novel type of deep radiomic features (DRFs) computed from a convolutional neural network (CNN), which capture tumor characteristics related to immune cell markers and overall survival. Our study uses four MRI sequences (T1-weighted, T1-weighted post-contrast, T2-weighted and FLAIR) with corresponding immune cell markers of 151 patients with brain tumor. The proposed method extracts a total of 180 DRFs by aggregating the activation maps of a pre-trained 3D-CNN within labeled tumor regions of MRI scans. These features offer a compact, yet powerful representation of regional texture encoding tissue heterogeneity. A comprehensive set of experiments is performed to assess the relationship between the proposed DRFs and immune cell markers, and measure their association with overall survival. Results show a high correlation between DRFs and various markers, as well as significant differences between patients grouped based on these markers. Moreover, combining DRFs, clinical features and immune cell markers as input to a random forest classifier helps discriminate between short and long survival outcomes, with AUC of 72\% and p=2.36$\times$10$^{-5}$. These results demonstrate the usefulness of proposed DRFs as non-invasive biomarker for predicting treatment response in patients with brain tumors.
Deep radiomic features from MRI scans predict survival outcome of recurrent glioblastoma
Nov 15, 2019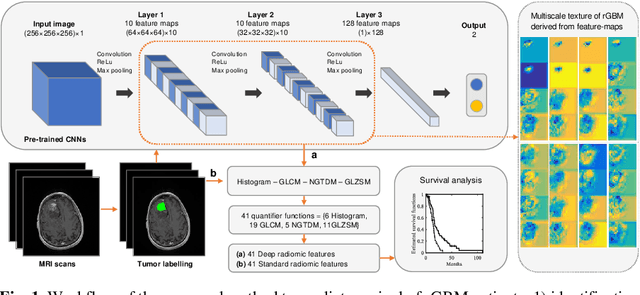
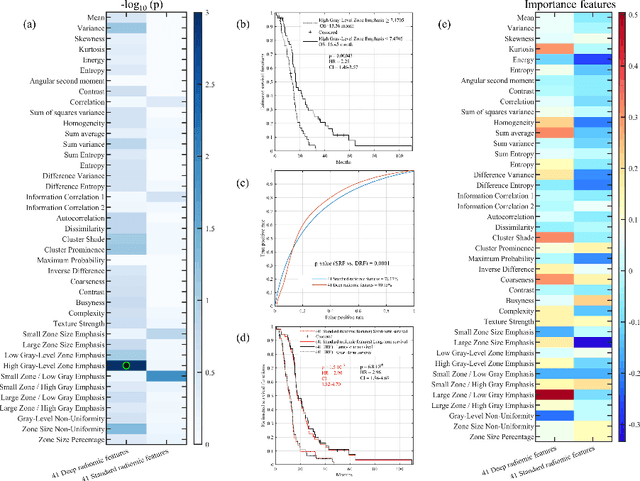
Abstract:This paper proposes to use deep radiomic features (DRFs) from a convolutional neural network (CNN) to model fine-grained texture signatures in the radiomic analysis of recurrent glioblastoma (rGBM). We use DRFs to predict survival of rGBM patients with preoperative T1-weighted post-contrast MR images (n=100). DRFs are extracted from regions of interest labelled by a radiation oncologist and used to compare between short-term and long-term survival patient groups. Random forest (RF) classification is employed to predict survival outcome (i.e., short or long survival), as well as to identify highly group-informative descriptors. Classification using DRFs results in an area under the ROC curve (AUC) of 89.15% (p<0.01) in predicting rGBM patient survival, compared to 78.07% (p<0.01) when using standard radiomic features (SRF). These results indicate the potential of DRFs as a prognostic marker for patients with rGBM.
Radiopathomics: Integration of radiographic and histologic characteristics for prognostication in glioblastoma
Sep 19, 2019



Abstract:Both radiographic (Rad) imaging, such as multi-parametric magnetic resonance imaging, and digital pathology (Path) images captured from tissue samples are currently acquired as standard clinical practice for glioblastoma tumors. Both these data streams have been separately used for diagnosis and treatment planning, despite the fact that they provide complementary information. In this research work, we aimed to assess the potential of both Rad and Path images in combination and comparison. An extensive set of engineered features was extracted from delineated tumor regions in Rad images, comprising T1, T1-Gd, T2, T2-FLAIR, and 100 random patches extracted from Path images. Specifically, the features comprised descriptors of intensity, histogram, and texture, mainly quantified via gray-level-co-occurrence matrix and gray-level-run-length matrices. Features extracted from images of 107 glioblastoma patients, downloaded from The Cancer Imaging Archive, were run through support vector machine for classification using leave-one-out cross-validation mechanism, and through support vector regression for prediction of continuous survival outcome. The Pearson correlation coefficient was estimated to be 0.75, 0.74, and 0.78 for Rad, Path and RadPath data. The area-under the receiver operating characteristic curve was estimated to be 0.74, 0.76 and 0.80 for Rad, Path and RadPath data, when patients were discretized into long- and short-survival groups based on average survival cutoff. Our results support the notion that synergistically using Rad and Path images may lead to better prognosis at the initial presentation of the disease, thereby facilitating the targeted enrollment of patients into clinical trials.
Prediction of overall survival and molecular markers in gliomas via analysis of digital pathology images using deep learning
Sep 19, 2019



Abstract:Cancer histology reveals disease progression and associated molecular processes, and contains rich phenotypic information that is predictive of outcome. In this paper, we developed a computational approach based on deep learning to predict the overall survival and molecular subtypes of glioma patients from microscopic images of tissue biopsies, reflecting measures of microvascular proliferation, mitotic activity, nuclear atypia, and the presence of necrosis. Whole-slide images from 663 unique patients [IDH: 333 IDH-wildtype, 330 IDH-mutants, 1p/19q: 201 1p/19q non-codeleted, 129 1p/19q codeleted] were obtained from TCGA. Sub-images that were free of artifacts and that contained viable tumor with descriptive histologic characteristics were extracted, which were further used for training and testing a deep neural network. The output layer of the network was configured in two different ways: (i) a final Cox model layer to output a prediction of patient risk, and (ii) a final layer with sigmoid activation function, and stochastic gradient decent based optimization with binary cross-entropy loss. Both survival prediction and molecular subtype classification produced promising results using our model. The c-statistic was estimated to be 0.82 (p-value=4.8x10-5) between the risk scores of the proposed deep learning model and overall survival, while accuracies of 88% (area under the curve [AUC]=0.86) were achieved in the detection of IDH mutational status and 1p/19q codeletion. These findings suggest that the deep learning techniques can be applied to microscopic images for objective, accurate, and integrated prediction of outcome for glioma patients. The proposed marker may contribute to (i) stratification of patients into clinical trials, (ii) patient selection for targeted therapy, and (iii) personalized treatment planning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge