Dominik Krzemiński
Cardiff University Brain Research Imaging Centre, Cardiff University, United Kingdom
Kaleidoscope: In-language Exams for Massively Multilingual Vision Evaluation
Apr 09, 2025Abstract:The evaluation of vision-language models (VLMs) has mainly relied on English-language benchmarks, leaving significant gaps in both multilingual and multicultural coverage. While multilingual benchmarks have expanded, both in size and languages, many rely on translations of English datasets, failing to capture cultural nuances. In this work, we propose Kaleidoscope, as the most comprehensive exam benchmark to date for the multilingual evaluation of vision-language models. Kaleidoscope is a large-scale, in-language multimodal benchmark designed to evaluate VLMs across diverse languages and visual inputs. Kaleidoscope covers 18 languages and 14 different subjects, amounting to a total of 20,911 multiple-choice questions. Built through an open science collaboration with a diverse group of researchers worldwide, Kaleidoscope ensures linguistic and cultural authenticity. We evaluate top-performing multilingual vision-language models and find that they perform poorly on low-resource languages and in complex multimodal scenarios. Our results highlight the need for progress on culturally inclusive multimodal evaluation frameworks.
MMTEB: Massive Multilingual Text Embedding Benchmark
Feb 19, 2025Abstract:Text embeddings are typically evaluated on a limited set of tasks, which are constrained by language, domain, and task diversity. To address these limitations and provide a more comprehensive evaluation, we introduce the Massive Multilingual Text Embedding Benchmark (MMTEB) - a large-scale, community-driven expansion of MTEB, covering over 500 quality-controlled evaluation tasks across 250+ languages. MMTEB includes a diverse set of challenging, novel tasks such as instruction following, long-document retrieval, and code retrieval, representing the largest multilingual collection of evaluation tasks for embedding models to date. Using this collection, we develop several highly multilingual benchmarks, which we use to evaluate a representative set of models. We find that while large language models (LLMs) with billions of parameters can achieve state-of-the-art performance on certain language subsets and task categories, the best-performing publicly available model is multilingual-e5-large-instruct with only 560 million parameters. To facilitate accessibility and reduce computational cost, we introduce a novel downsampling method based on inter-task correlation, ensuring a diverse selection while preserving relative model rankings. Furthermore, we optimize tasks such as retrieval by sampling hard negatives, creating smaller but effective splits. These optimizations allow us to introduce benchmarks that drastically reduce computational demands. For instance, our newly introduced zero-shot English benchmark maintains a ranking order similar to the full-scale version but at a fraction of the computational cost.
INCLUDE: Evaluating Multilingual Language Understanding with Regional Knowledge
Nov 29, 2024



Abstract:The performance differential of large language models (LLM) between languages hinders their effective deployment in many regions, inhibiting the potential economic and societal value of generative AI tools in many communities. However, the development of functional LLMs in many languages (\ie, multilingual LLMs) is bottlenecked by the lack of high-quality evaluation resources in languages other than English. Moreover, current practices in multilingual benchmark construction often translate English resources, ignoring the regional and cultural knowledge of the environments in which multilingual systems would be used. In this work, we construct an evaluation suite of 197,243 QA pairs from local exam sources to measure the capabilities of multilingual LLMs in a variety of regional contexts. Our novel resource, INCLUDE, is a comprehensive knowledge- and reasoning-centric benchmark across 44 written languages that evaluates multilingual LLMs for performance in the actual language environments where they would be deployed.
Aya Dataset: An Open-Access Collection for Multilingual Instruction Tuning
Feb 09, 2024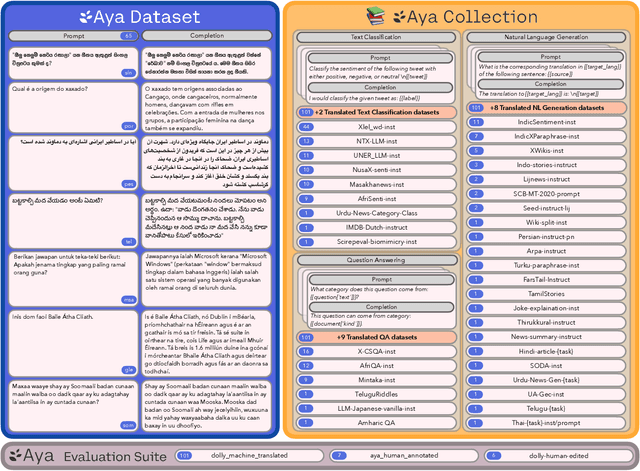
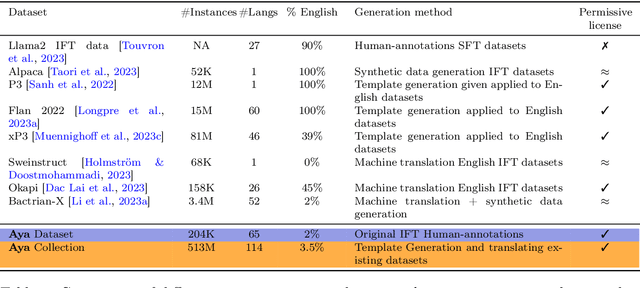
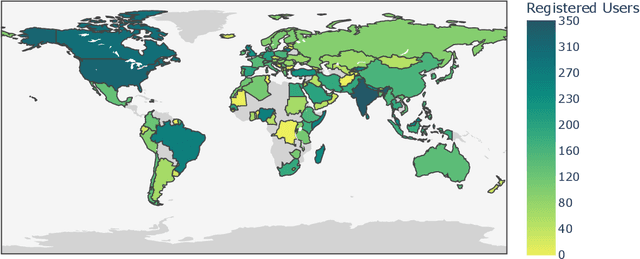
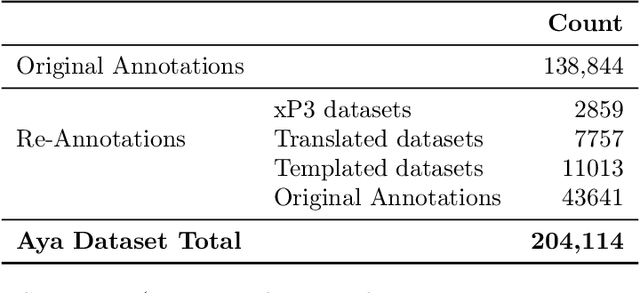
Abstract:Datasets are foundational to many breakthroughs in modern artificial intelligence. Many recent achievements in the space of natural language processing (NLP) can be attributed to the finetuning of pre-trained models on a diverse set of tasks that enables a large language model (LLM) to respond to instructions. Instruction fine-tuning (IFT) requires specifically constructed and annotated datasets. However, existing datasets are almost all in the English language. In this work, our primary goal is to bridge the language gap by building a human-curated instruction-following dataset spanning 65 languages. We worked with fluent speakers of languages from around the world to collect natural instances of instructions and completions. Furthermore, we create the most extensive multilingual collection to date, comprising 513 million instances through templating and translating existing datasets across 114 languages. In total, we contribute four key resources: we develop and open-source the Aya Annotation Platform, the Aya Dataset, the Aya Collection, and the Aya Evaluation Suite. The Aya initiative also serves as a valuable case study in participatory research, involving collaborators from 119 countries. We see this as a valuable framework for future research collaborations that aim to bridge gaps in resources.
Open source software for automatic subregional assessment of knee cartilage degradation using quantitative T2 relaxometry and deep learning
Dec 22, 2020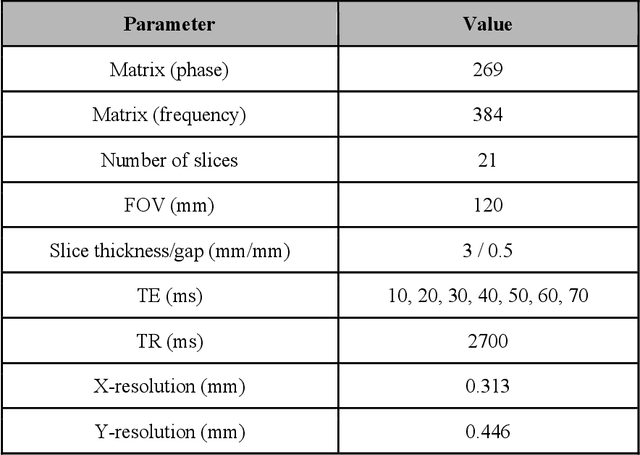
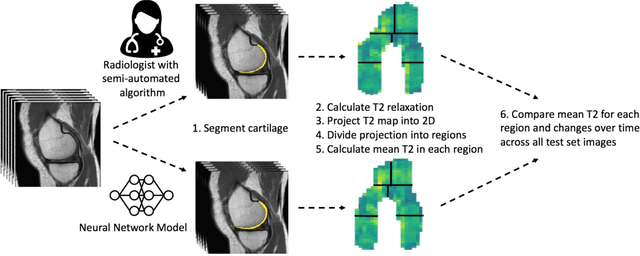
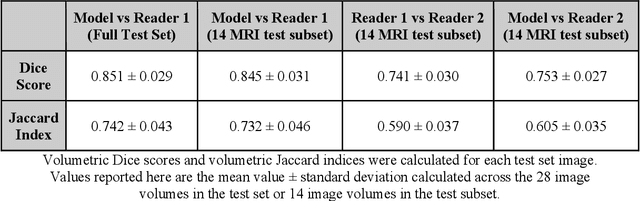
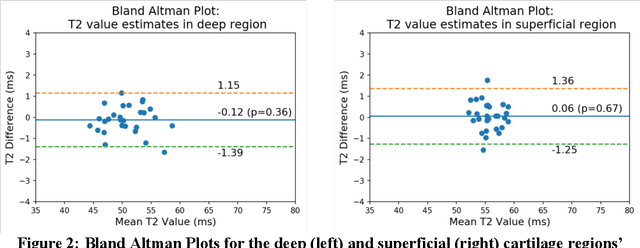
Abstract:Objective: We evaluate a fully-automated femoral cartilage segmentation model for measuring T2 relaxation values and longitudinal changes using multi-echo spin echo (MESE) MRI. We have open sourced this model and corresponding segmentations. Methods: We trained a neural network to segment femoral cartilage from MESE MRIs. Cartilage was divided into 12 subregions along medial-lateral, superficial-deep, and anterior-central-posterior boundaries. Subregional T2 values and four-year changes were calculated using a musculoskeletal radiologist's segmentations (Reader 1) and the model's segmentations. These were compared using 28 held out images. A subset of 14 images were also evaluated by a second expert (Reader 2) for comparison. Results: Model segmentations agreed with Reader 1 segmentations with a Dice score of 0.85 +/- 0.03. The model's estimated T2 values for individual subregions agreed with those of Reader 1 with an average Spearman correlation of 0.89 and average mean absolute error (MAE) of 1.34 ms. The model's estimated four-year change in T2 for individual regions agreed with Reader 1 with an average correlation of 0.80 and average MAE of 1.72 ms. The model agreed with Reader 1 at least as closely as Reader 2 agreed with Reader 1 in terms of Dice score (0.85 vs 0.75) and subregional T2 values. Conclusions: We present a fast, fully-automated model for segmentation of MESE MRIs. Assessments of cartilage health using its segmentations agree with those of an expert as closely as experts agree with one another. This has the potential to accelerate osteoarthritis research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge