Garry E. Gold
Department of Biomedical Engineering, Stanford University, California, USA, Department of Radiology, Stanford University, California, USA, Department of Orthopaedic Surgery, Stanford University, California, USA
Unsupervised Training of a Dynamic Context-Aware Deep Denoising Framework for Low-Dose Fluoroscopic Imaging
Oct 29, 2024



Abstract:Fluoroscopy is critical for real-time X-ray visualization in medical imaging. However, low-dose images are compromised by noise, potentially affecting diagnostic accuracy. Noise reduction is crucial for maintaining image quality, especially given such challenges as motion artifacts and the limited availability of clean data in medical imaging. To address these issues, we propose an unsupervised training framework for dynamic context-aware denoising of fluoroscopy image sequences. First, we train the multi-scale recurrent attention U-Net (MSR2AU-Net) without requiring clean data to address the initial noise. Second, we incorporate a knowledge distillation-based uncorrelated noise suppression module and a recursive filtering-based correlated noise suppression module enhanced with motion compensation to further improve motion compensation and achieve superior denoising performance. Finally, we introduce a novel approach by combining these modules with a pixel-wise dynamic object motion cross-fusion matrix, designed to adapt to motion, and an edge-preserving loss for precise detail retention. To validate the proposed method, we conducted extensive numerical experiments on medical image datasets, including 3500 fluoroscopy images from dynamic phantoms (2,400 images for training, 1,100 for testing) and 350 clinical images from a spinal surgery patient. Moreover, we demonstrated the robustness of our approach across different imaging modalities by testing it on the publicly available 2016 Low Dose CT Grand Challenge dataset, using 4,800 images for training and 1,136 for testing. The results demonstrate that the proposed approach outperforms state-of-the-art unsupervised algorithms in both visual quality and quantitative evaluation while achieving comparable performance to well-established supervised learning methods across low-dose fluoroscopy and CT imaging.
Data-Limited Tissue Segmentation using Inpainting-Based Self-Supervised Learning
Oct 14, 2022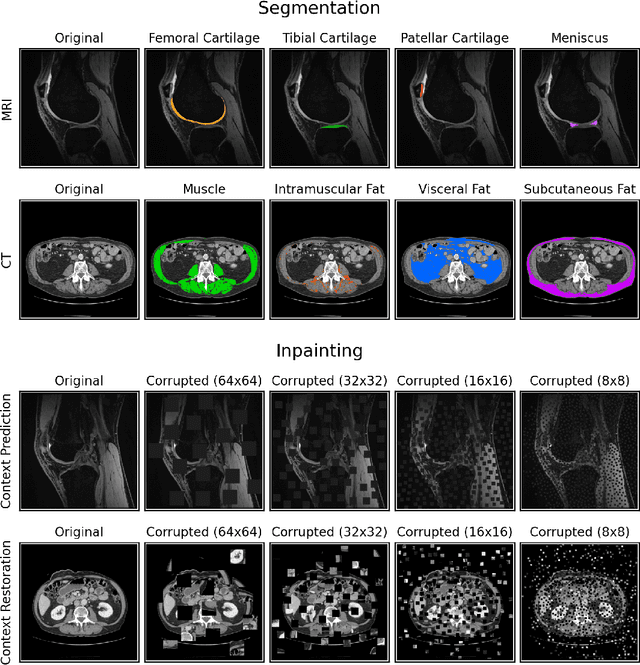
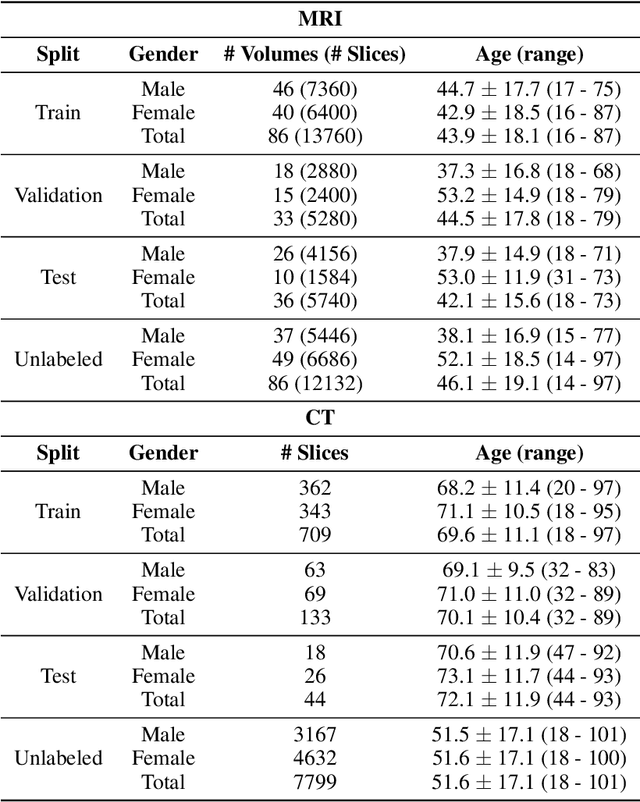
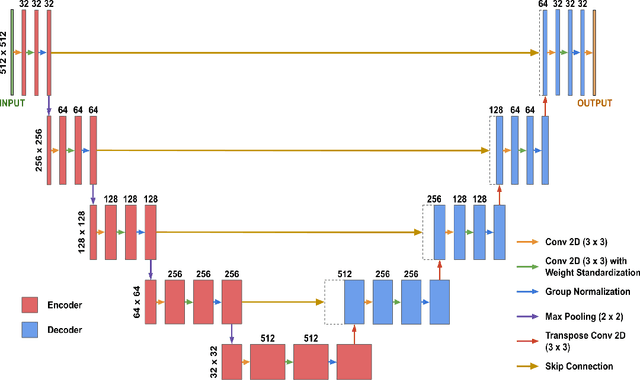
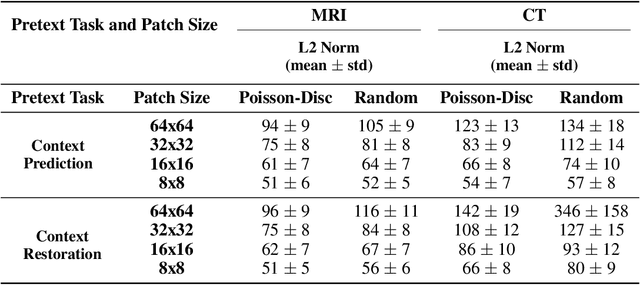
Abstract:Although supervised learning has enabled high performance for image segmentation, it requires a large amount of labeled training data, which can be difficult to obtain in the medical imaging field. Self-supervised learning (SSL) methods involving pretext tasks have shown promise in overcoming this requirement by first pretraining models using unlabeled data. In this work, we evaluate the efficacy of two SSL methods (inpainting-based pretext tasks of context prediction and context restoration) for CT and MRI image segmentation in label-limited scenarios, and investigate the effect of implementation design choices for SSL on downstream segmentation performance. We demonstrate that optimally trained and easy-to-implement inpainting-based SSL segmentation models can outperform classically supervised methods for MRI and CT tissue segmentation in label-limited scenarios, for both clinically-relevant metrics and the traditional Dice score.
Open source software for automatic subregional assessment of knee cartilage degradation using quantitative T2 relaxometry and deep learning
Dec 22, 2020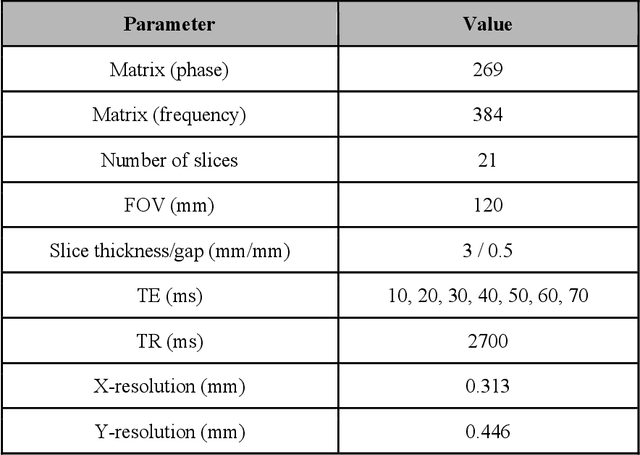
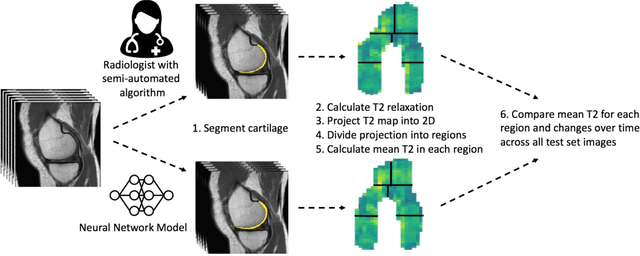
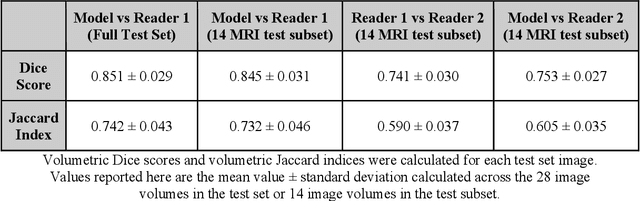
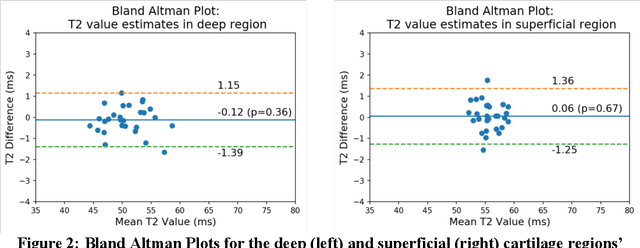
Abstract:Objective: We evaluate a fully-automated femoral cartilage segmentation model for measuring T2 relaxation values and longitudinal changes using multi-echo spin echo (MESE) MRI. We have open sourced this model and corresponding segmentations. Methods: We trained a neural network to segment femoral cartilage from MESE MRIs. Cartilage was divided into 12 subregions along medial-lateral, superficial-deep, and anterior-central-posterior boundaries. Subregional T2 values and four-year changes were calculated using a musculoskeletal radiologist's segmentations (Reader 1) and the model's segmentations. These were compared using 28 held out images. A subset of 14 images were also evaluated by a second expert (Reader 2) for comparison. Results: Model segmentations agreed with Reader 1 segmentations with a Dice score of 0.85 +/- 0.03. The model's estimated T2 values for individual subregions agreed with those of Reader 1 with an average Spearman correlation of 0.89 and average mean absolute error (MAE) of 1.34 ms. The model's estimated four-year change in T2 for individual regions agreed with Reader 1 with an average correlation of 0.80 and average MAE of 1.72 ms. The model agreed with Reader 1 at least as closely as Reader 2 agreed with Reader 1 in terms of Dice score (0.85 vs 0.75) and subregional T2 values. Conclusions: We present a fast, fully-automated model for segmentation of MESE MRIs. Assessments of cartilage health using its segmentations agree with those of an expert as closely as experts agree with one another. This has the potential to accelerate osteoarthritis research.
The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset
May 26, 2020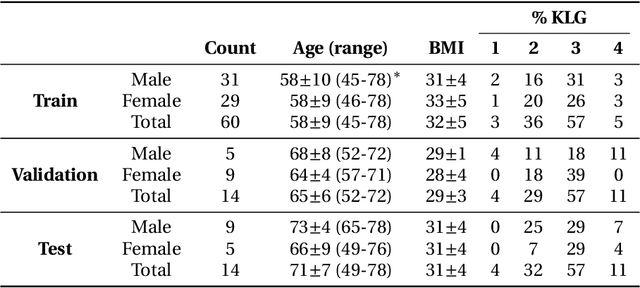
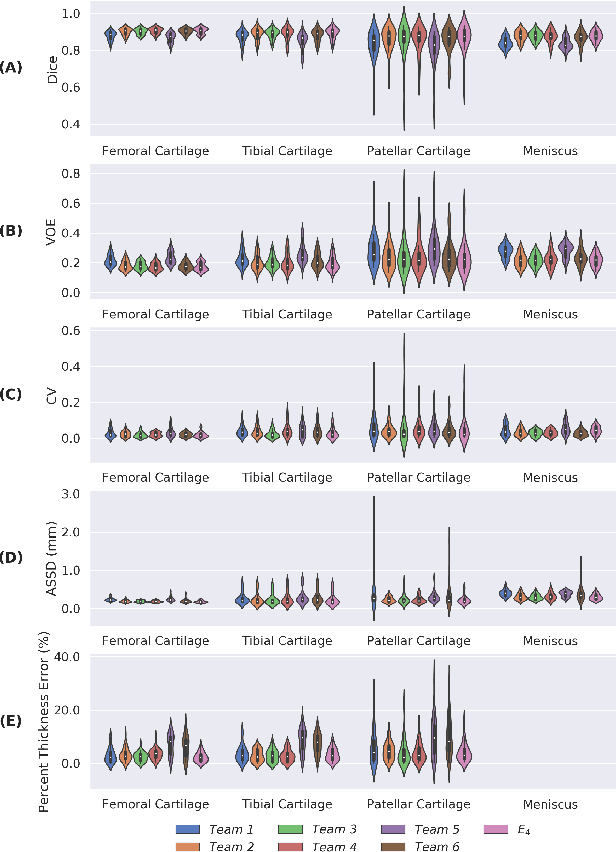
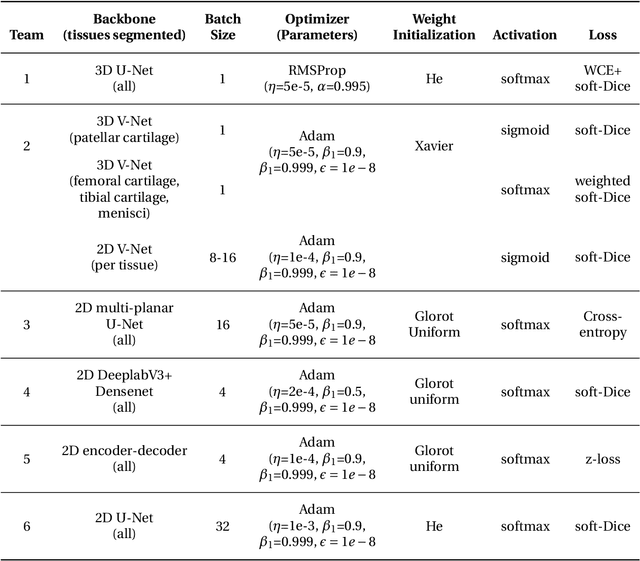
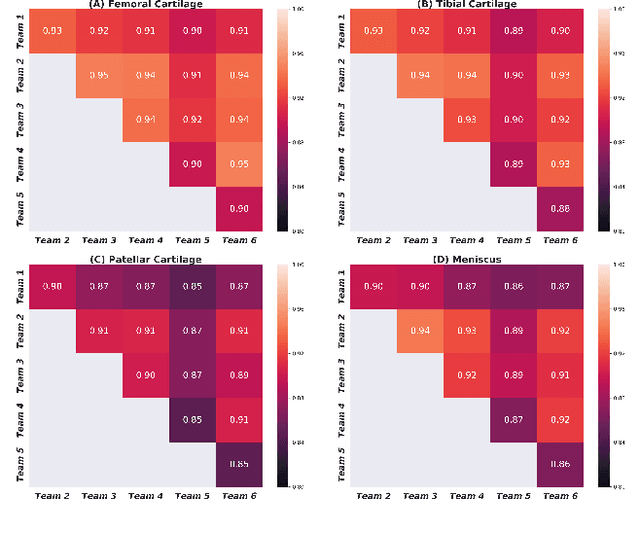
Abstract:Purpose: To organize a knee MRI segmentation challenge for characterizing the semantic and clinical efficacy of automatic segmentation methods relevant for monitoring osteoarthritis progression. Methods: A dataset partition consisting of 3D knee MRI from 88 subjects at two timepoints with ground-truth articular (femoral, tibial, patellar) cartilage and meniscus segmentations was standardized. Challenge submissions and a majority-vote ensemble were evaluated using Dice score, average symmetric surface distance, volumetric overlap error, and coefficient of variation on a hold-out test set. Similarities in network segmentations were evaluated using pairwise Dice correlations. Articular cartilage thickness was computed per-scan and longitudinally. Correlation between thickness error and segmentation metrics was measured using Pearson's coefficient. Two empirical upper bounds for ensemble performance were computed using combinations of model outputs that consolidated true positives and true negatives. Results: Six teams (T1-T6) submitted entries for the challenge. No significant differences were observed across all segmentation metrics for all tissues (p=1.0) among the four top-performing networks (T2, T3, T4, T6). Dice correlations between network pairs were high (>0.85). Per-scan thickness errors were negligible among T1-T4 (p=0.99) and longitudinal changes showed minimal bias (<0.03mm). Low correlations (<0.41) were observed between segmentation metrics and thickness error. The majority-vote ensemble was comparable to top performing networks (p=1.0). Empirical upper bound performances were similar for both combinations (p=1.0). Conclusion: Diverse networks learned to segment the knee similarly where high segmentation accuracy did not correlate to cartilage thickness accuracy. Voting ensembles did not outperform individual networks but may help regularize individual models.
Technical Considerations for Semantic Segmentation in MRI using Convolutional Neural Networks
Feb 05, 2019



Abstract:High-fidelity semantic segmentation of magnetic resonance volumes is critical for estimating tissue morphometry and relaxation parameters in both clinical and research applications. While manual segmentation is accepted as the gold-standard, recent advances in deep learning and convolutional neural networks (CNNs) have shown promise for efficient automatic segmentation of soft tissues. However, due to the stochastic nature of deep learning and the multitude of hyperparameters in training networks, predicting network behavior is challenging. In this paper, we quantify the impact of three factors associated with CNN segmentation performance: network architecture, training loss functions, and training data characteristics. We evaluate the impact of these variations on the segmentation of femoral cartilage and propose potential modifications to CNN architectures and training protocols to train these models with confidence.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge