Claudia Iriondo
Counterfactual Generative Modeling with Variational Causal Inference
Oct 16, 2024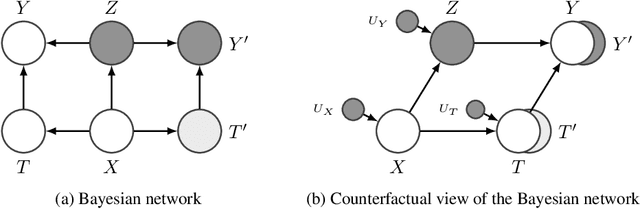
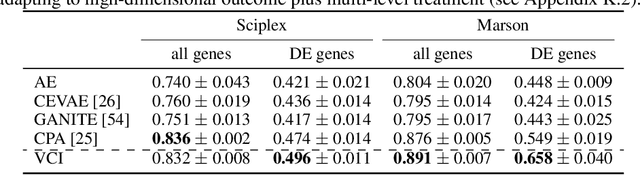
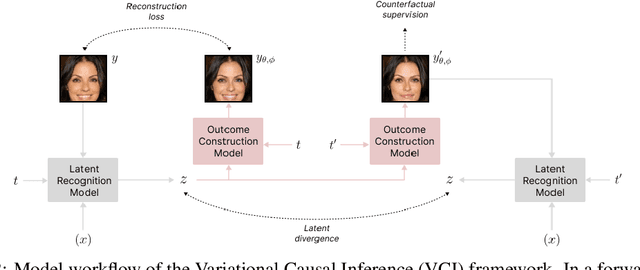
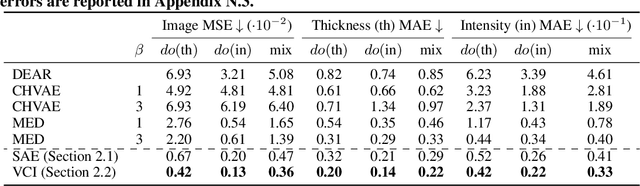
Abstract:Estimating an individual's potential outcomes under counterfactual treatments is a challenging task for traditional causal inference and supervised learning approaches when the outcome is high-dimensional (e.g. gene expressions, facial images) and covariates are relatively limited. In this case, to predict one's outcomes under counterfactual treatments, it is crucial to leverage individual information contained in its high-dimensional observed outcome in addition to the covariates. Prior works using variational inference in counterfactual generative modeling have been focusing on neural adaptations and model variants within the conditional variational autoencoder formulation, which we argue is fundamentally ill-suited to the notion of counterfactual in causal inference. In this work, we present a novel variational Bayesian causal inference framework and its theoretical backings to properly handle counterfactual generative modeling tasks, through which we are able to conduct counterfactual supervision end-to-end during training without any counterfactual samples, and encourage latent disentanglement that aids the correct identification of causal effect in counterfactual generations. In experiments, we demonstrate the advantage of our framework compared to state-of-the-art models in counterfactual generative modeling on multiple benchmarks.
DeepAD: A Robust Deep Learning Model of Alzheimer's Disease Progression for Real-World Clinical Applications
Apr 08, 2022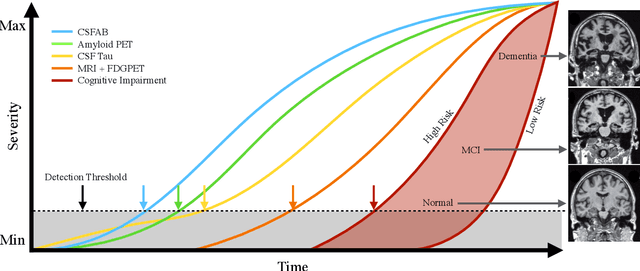
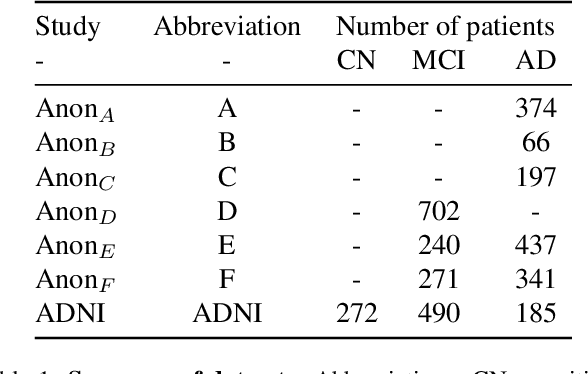
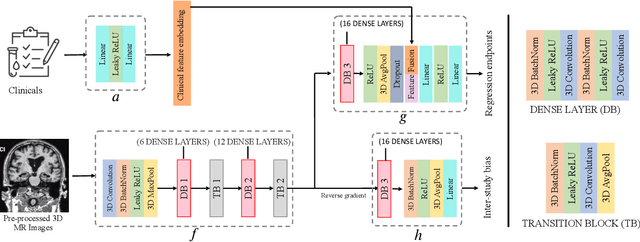
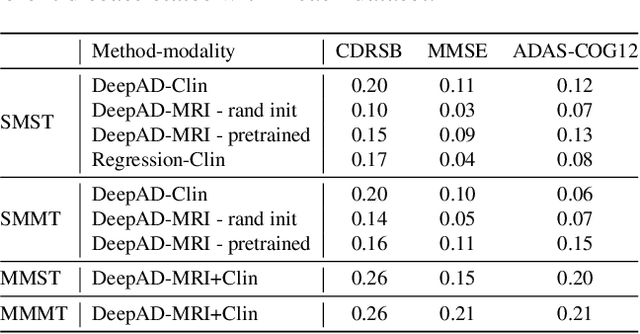
Abstract:The ability to predict the future trajectory of a patient is a key step toward the development of therapeutics for complex diseases such as Alzheimer's disease (AD). However, most machine learning approaches developed for prediction of disease progression are either single-task or single-modality models, which can not be directly adopted to our setting involving multi-task learning with high dimensional images. Moreover, most of those approaches are trained on a single dataset (i.e. cohort), which can not be generalized to other cohorts. We propose a novel multimodal multi-task deep learning model to predict AD progression by analyzing longitudinal clinical and neuroimaging data from multiple cohorts. Our proposed model integrates high dimensional MRI features from a 3D convolutional neural network with other data modalities, including clinical and demographic information, to predict the future trajectory of patients. Our model employs an adversarial loss to alleviate the study-specific imaging bias, in particular the inter-study domain shifts. In addition, a Sharpness-Aware Minimization (SAM) optimization technique is applied to further improve model generalization. The proposed model is trained and tested on various datasets in order to evaluate and validate the results. Our results showed that 1) our model yields significant improvement over the baseline models, and 2) models using extracted neuroimaging features from 3D convolutional neural network outperform the same models when applied to MRI-derived volumetric features.
The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset
May 26, 2020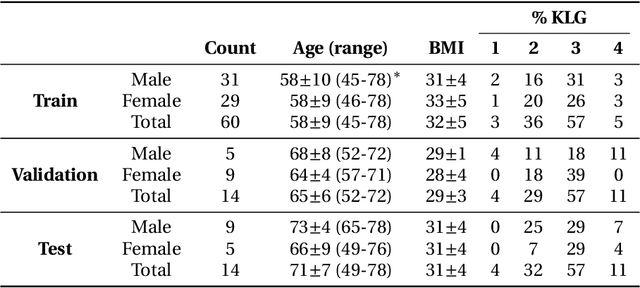
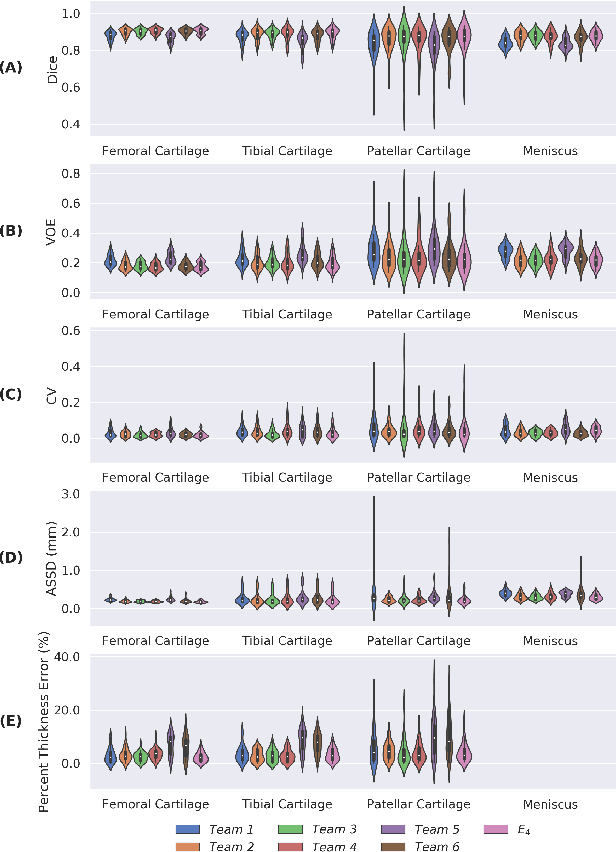
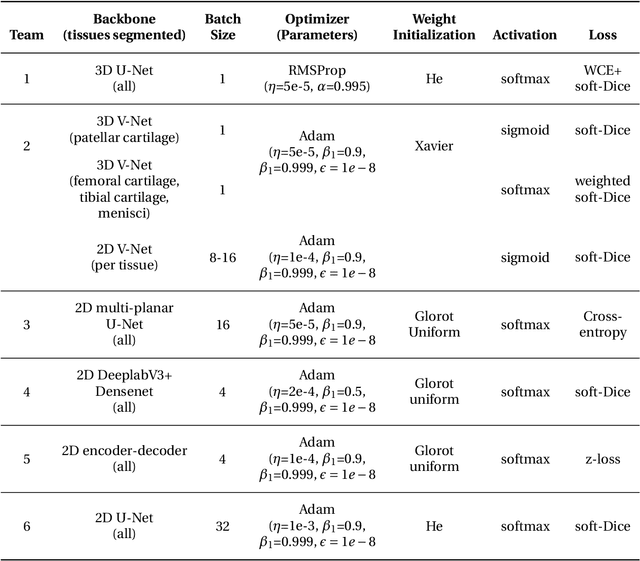
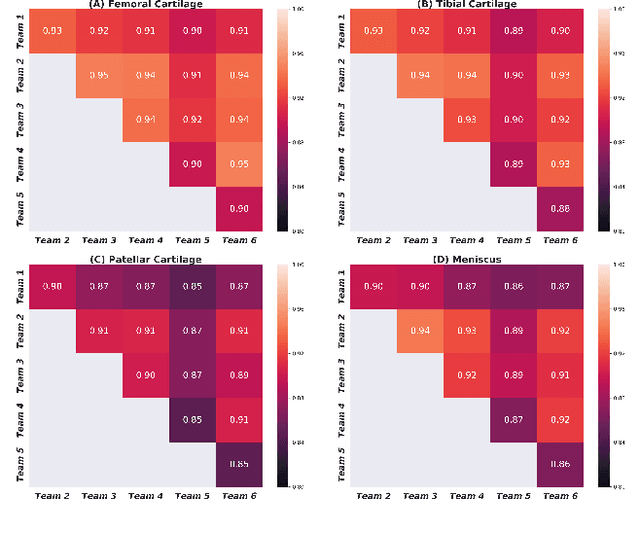
Abstract:Purpose: To organize a knee MRI segmentation challenge for characterizing the semantic and clinical efficacy of automatic segmentation methods relevant for monitoring osteoarthritis progression. Methods: A dataset partition consisting of 3D knee MRI from 88 subjects at two timepoints with ground-truth articular (femoral, tibial, patellar) cartilage and meniscus segmentations was standardized. Challenge submissions and a majority-vote ensemble were evaluated using Dice score, average symmetric surface distance, volumetric overlap error, and coefficient of variation on a hold-out test set. Similarities in network segmentations were evaluated using pairwise Dice correlations. Articular cartilage thickness was computed per-scan and longitudinally. Correlation between thickness error and segmentation metrics was measured using Pearson's coefficient. Two empirical upper bounds for ensemble performance were computed using combinations of model outputs that consolidated true positives and true negatives. Results: Six teams (T1-T6) submitted entries for the challenge. No significant differences were observed across all segmentation metrics for all tissues (p=1.0) among the four top-performing networks (T2, T3, T4, T6). Dice correlations between network pairs were high (>0.85). Per-scan thickness errors were negligible among T1-T4 (p=0.99) and longitudinal changes showed minimal bias (<0.03mm). Low correlations (<0.41) were observed between segmentation metrics and thickness error. The majority-vote ensemble was comparable to top performing networks (p=1.0). Empirical upper bound performances were similar for both combinations (p=1.0). Conclusion: Diverse networks learned to segment the knee similarly where high segmentation accuracy did not correlate to cartilage thickness accuracy. Voting ensembles did not outperform individual networks but may help regularize individual models.
Adversarial Policy Gradient for Deep Learning Image Augmentation
Sep 09, 2019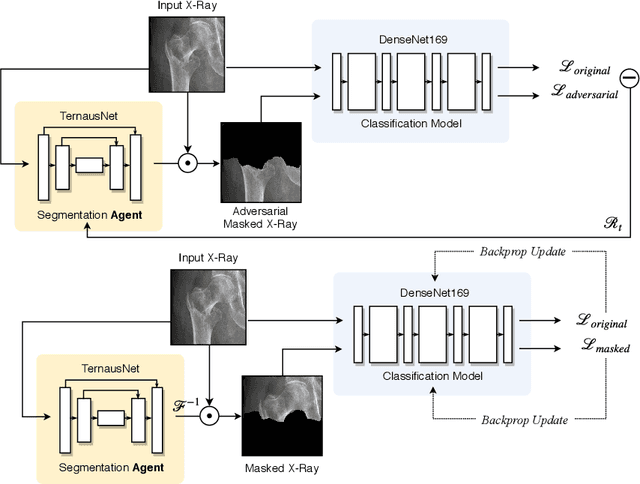
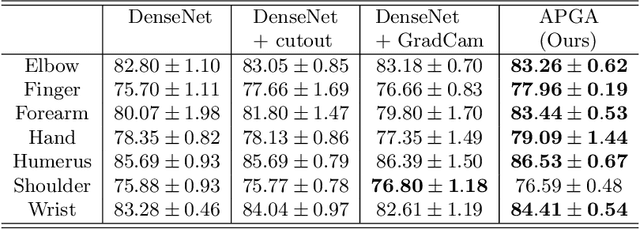
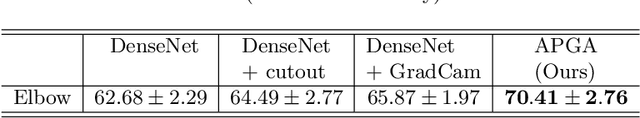
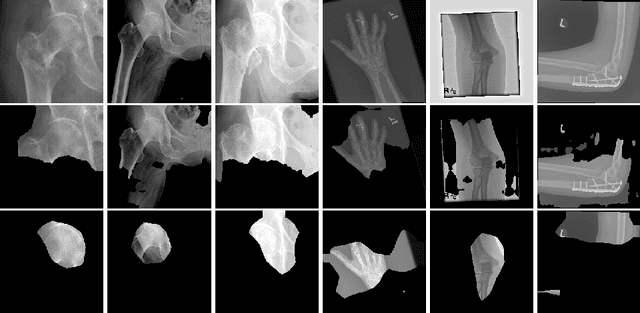
Abstract:The use of semantic segmentation for masking and cropping input images has proven to be a significant aid in medical imaging classification tasks by decreasing the noise and variance of the training dataset. However, implementing this approach with classical methods is challenging: the cost of obtaining a dense segmentation is high, and the precise input area that is most crucial to the classification task is difficult to determine a-priori. We propose a novel joint-training deep reinforcement learning framework for image augmentation. A segmentation network, weakly supervised with policy gradient optimization, acts as an agent, and outputs masks as actions given samples as states, with the goal of maximizing reward signals from the classification network. In this way, the segmentation network learns to mask unimportant imaging features. Our method, Adversarial Policy Gradient Augmentation (APGA), shows promising results on Stanford's MURA dataset and on a hip fracture classification task with an increase in global accuracy of up to 7.33% and improved performance over baseline methods in 9/10 tasks evaluated. We discuss the broad applicability of our joint training strategy to a variety of medical imaging tasks.
Distance Map Loss Penalty Term for Semantic Segmentation
Aug 10, 2019
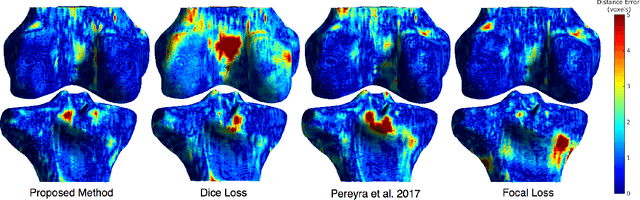
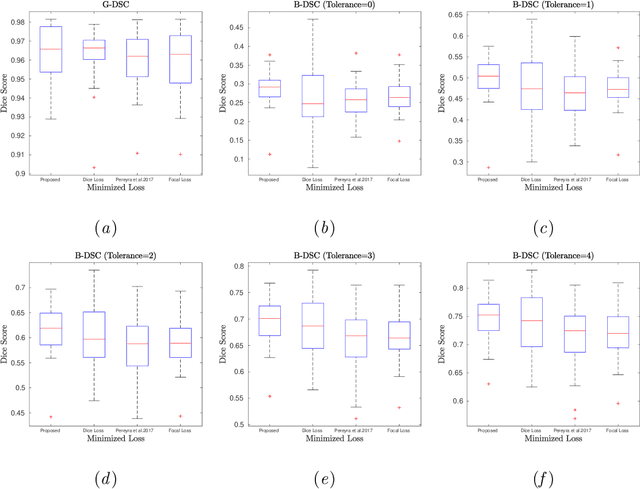
Abstract:Convolutional neural networks for semantic segmentation suffer from low performance at object boundaries. In medical imaging, accurate representation of tissue surfaces and volumes is important for tracking of disease biomarkers such as tissue morphology and shape features. In this work, we propose a novel distance map derived loss penalty term for semantic segmentation. We propose to use distance maps, derived from ground truth masks, to create a penalty term, guiding the network's focus towards hard-to-segment boundary regions. We investigate the effects of this penalizing factor against cross-entropy, Dice, and focal loss, among others, evaluating performance on a 3D MRI bone segmentation task from the publicly available Osteoarthritis Initiative dataset. We observe a significant improvement in the quality of segmentation, with better shape preservation at bone boundaries and areas affected by partial volume. We ultimately aim to use our loss penalty term to improve the extraction of shape biomarkers and derive metrics to quantitatively evaluate the preservation of shape.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge