Somaye Hashemifar
Speaking the Same Language: Leveraging LLMs in Standardizing Clinical Data for AI
Aug 16, 2024Abstract:The implementation of Artificial Intelligence (AI) in the healthcare industry has garnered considerable attention, attributable to its prospective enhancement of clinical outcomes, expansion of access to superior healthcare, cost reduction, and elevation of patient satisfaction. Nevertheless, the primary hurdle that persists is related to the quality of accessible multi-modal healthcare data in conjunction with the evolution of AI methodologies. This study delves into the adoption of large language models to address specific challenges, specifically, the standardization of healthcare data. We advocate the use of these models to identify and map clinical data schemas to established data standard attributes, such as the Fast Healthcare Interoperability Resources. Our results illustrate that employing large language models significantly diminishes the necessity for manual data curation and elevates the efficacy of the data standardization process. Consequently, the proposed methodology has the propensity to expedite the integration of AI in healthcare, ameliorate the quality of patient care, whilst minimizing the time and financial resources necessary for the preparation of data for AI.
Self-supervised Learning for Segmentation and Quantification of Dopamine Neurons in $\text{Parkinson's Disease}$
Jan 11, 2023Abstract:$\text{Parkinson's Disease}$ (PD) is the second most common neurodegenerative disease in humans. PD is characterized by the gradual loss of dopaminergic neurons in the Substantia Nigra (a part of the mid-brain). Counting the number of dopaminergic neurons in the Substantia Nigra is one of the most important indexes in evaluating drug efficacy in PD animal models. Currently, analyzing and quantifying dopaminergic neurons is conducted manually by experts through analysis of digital pathology images which is laborious, time-consuming, and highly subjective. As such, a reliable and unbiased automated system is demanded for the quantification of dopaminergic neurons in digital pathology images. We propose an end-to-end deep learning framework for the segmentation and quantification of dopaminergic neurons in PD animal models. To the best of knowledge, this is the first machine learning model that detects the cell body of dopaminergic neurons, counts the number of dopaminergic neurons and provides the phenotypic characteristics of individual dopaminergic neurons as a numerical output. Extensive experiments demonstrate the effectiveness of our model in quantifying neurons with a high precision, which can provide quicker turnaround for drug efficacy studies, better understanding of dopaminergic neuronal health status and unbiased results in PD pre-clinical research.
Multiclass Semantic Segmentation to Identify Anatomical Sub-Regions of Brain and Measure Neuronal Health in Parkinson's Disease
Jan 07, 2023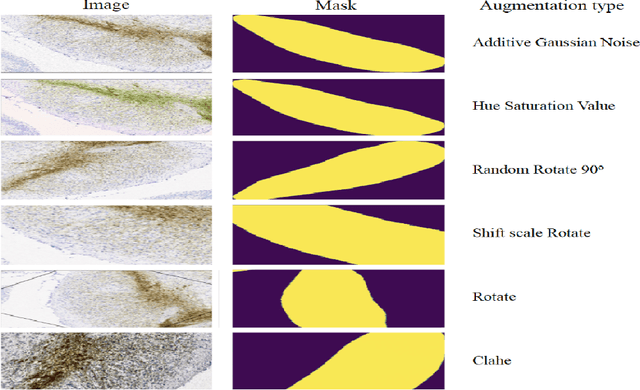
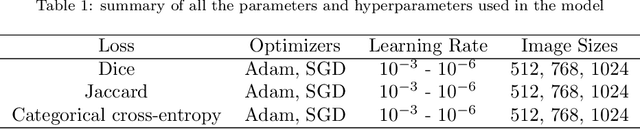
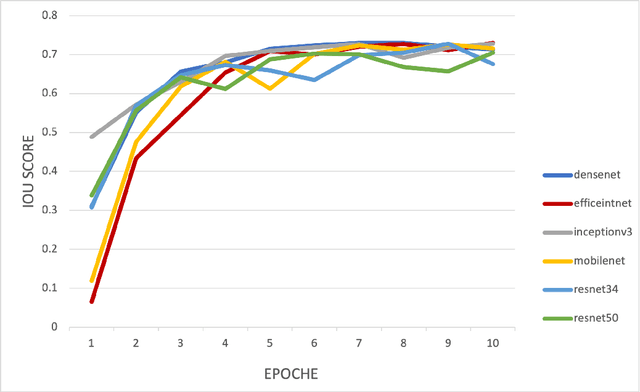

Abstract:Automated segmentation of anatomical sub-regions with high precision has become a necessity to enable the quantification and characterization of cells/ tissues in histology images. Currently, a machine learning model to analyze sub-anatomical regions of the brain to analyze 2D histological images is not available. The scientists rely on manually segmenting anatomical sub-regions of the brain which is extremely time-consuming and prone to labeler-dependent bias. One of the major challenges in accomplishing such a task is the lack of high-quality annotated images that can be used to train a generic artificial intelligence model. In this study, we employed a UNet-based architecture, compared model performance with various combinations of encoders, image sizes, and sample selection techniques. Additionally, to increase the sample set we resorted to data augmentation which provided data diversity and robust learning. In this study, we trained our best fit model on approximately one thousand annotated 2D brain images stained with Nissl/ Haematoxylin and Tyrosine Hydroxylase enzyme (TH, indicator of dopaminergic neuron viability). The dataset comprises of different animal studies enabling the model to be trained on different datasets. The model effectively is able to detect two sub-regions compacta (SNCD) and reticulata (SNr) in all the images. In spite of limited training data, our best model achieves a mean intersection over union (IOU) of 79% and a mean dice coefficient of 87%. In conclusion, the UNet-based model with EffiecientNet as an encoder outperforms all other encoders, resulting in a first of its kind robust model for multiclass segmentation of sub-brain regions in 2D images.
Cross-Domain Self-Supervised Deep Learning for Robust Alzheimer's Disease Progression Modeling
Nov 15, 2022Abstract:Developing successful artificial intelligence systems in practice depends both on robust deep learning models as well as large high quality data. Acquiring and labeling data can become prohibitively expensive and time-consuming in many real-world applications such as clinical disease models. Self-supervised learning has demonstrated great potential in increasing model accuracy and robustness in small data regimes. In addition, many clinical imaging and disease modeling applications rely heavily on regression of continuous quantities. However, the applicability of self-supervised learning for these medical-imaging regression tasks has not been extensively studied. In this study, we develop a cross-domain self-supervised learning approach for disease prognostic modeling as a regression problem using 3D images as input. We demonstrate that self-supervised pre-training can improve the prediction of Alzheimer's Disease progression from brain MRI. We also show that pre-training on extended (but not labeled) brain MRI data outperforms pre-training on natural images. We further observe that the highest performance is achieved when both natural images and extended brain-MRI data are used for pre-training.
DeepAD: A Robust Deep Learning Model of Alzheimer's Disease Progression for Real-World Clinical Applications
Apr 08, 2022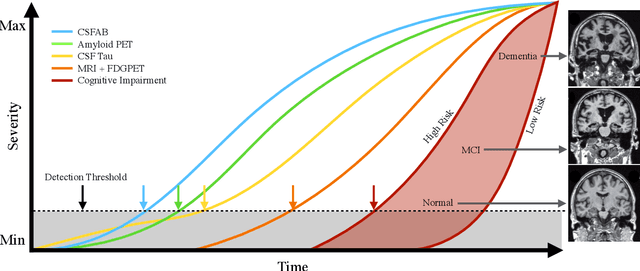
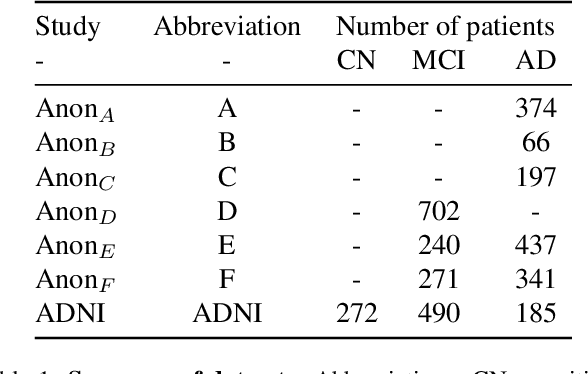
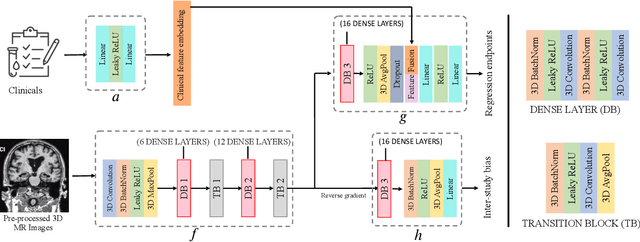
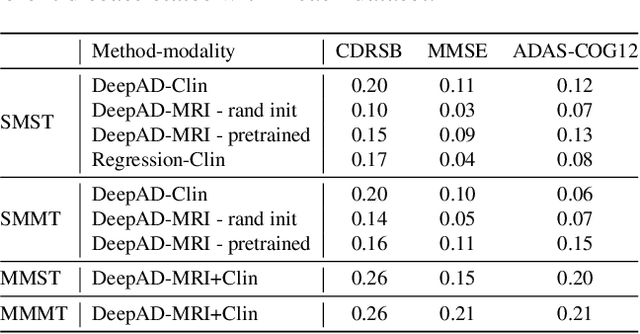
Abstract:The ability to predict the future trajectory of a patient is a key step toward the development of therapeutics for complex diseases such as Alzheimer's disease (AD). However, most machine learning approaches developed for prediction of disease progression are either single-task or single-modality models, which can not be directly adopted to our setting involving multi-task learning with high dimensional images. Moreover, most of those approaches are trained on a single dataset (i.e. cohort), which can not be generalized to other cohorts. We propose a novel multimodal multi-task deep learning model to predict AD progression by analyzing longitudinal clinical and neuroimaging data from multiple cohorts. Our proposed model integrates high dimensional MRI features from a 3D convolutional neural network with other data modalities, including clinical and demographic information, to predict the future trajectory of patients. Our model employs an adversarial loss to alleviate the study-specific imaging bias, in particular the inter-study domain shifts. In addition, a Sharpness-Aware Minimization (SAM) optimization technique is applied to further improve model generalization. The proposed model is trained and tested on various datasets in order to evaluate and validate the results. Our results showed that 1) our model yields significant improvement over the baseline models, and 2) models using extracted neuroimaging features from 3D convolutional neural network outperform the same models when applied to MRI-derived volumetric features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge