Hosein Barzekar
CELI: Controller-Embedded Language Model Interactions
Oct 18, 2024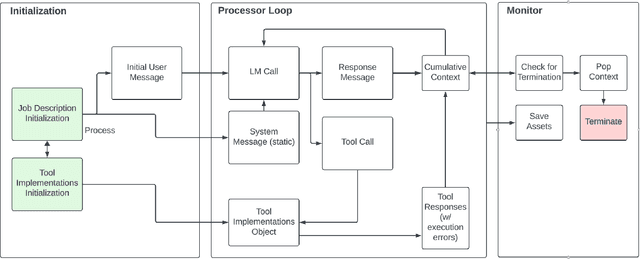
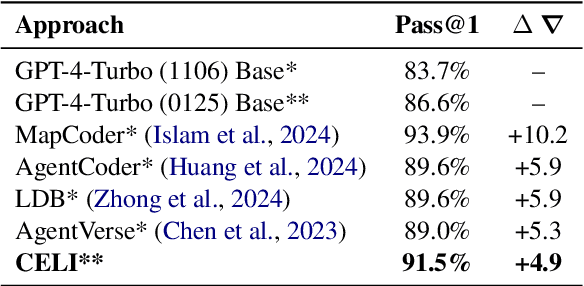
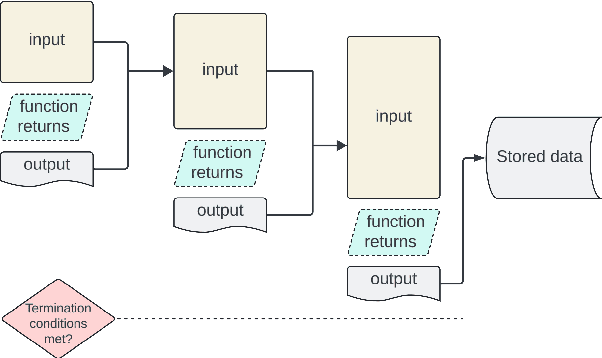
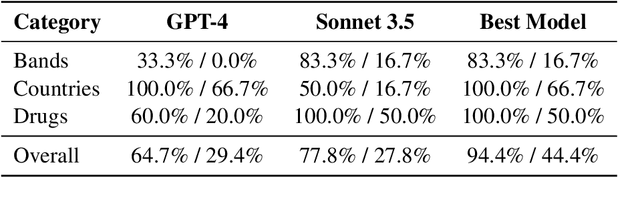
Abstract:We introduce Controller-Embedded Language Model Interactions (CELI), a framework that integrates control logic directly within language model (LM) prompts, facilitating complex, multi-stage task execution. CELI addresses limitations of existing prompt engineering and workflow optimization techniques by embedding control logic directly within the operational context of language models, enabling dynamic adaptation to evolving task requirements. Our framework transfers control from the traditional programming execution environment to the LMs, allowing them to autonomously manage computational workflows while maintaining seamless interaction with external systems and functions. CELI supports arbitrary function calls with variable arguments, bridging the gap between LMs' adaptive reasoning capabilities and conventional software paradigms' structured control mechanisms. To evaluate CELI's versatility and effectiveness, we conducted case studies in two distinct domains: code generation (HumanEval benchmark) and multi-stage content generation (Wikipedia-style articles). The results demonstrate notable performance improvements across a range of domains. CELI achieved a 4.9 percentage point improvement over the best reported score of the baseline GPT-4 model on the HumanEval code generation benchmark. In multi-stage content generation, 94.4% of CELI-produced Wikipedia-style articles met or exceeded first draft quality when optimally configured, with 44.4% achieving high quality. These outcomes underscore CELI's potential for optimizing AI-driven workflows across diverse computational domains.
MultiNet with Transformers: A Model for Cancer Diagnosis Using Images
Jan 21, 2023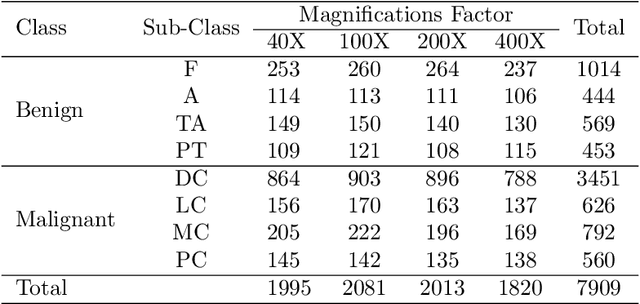
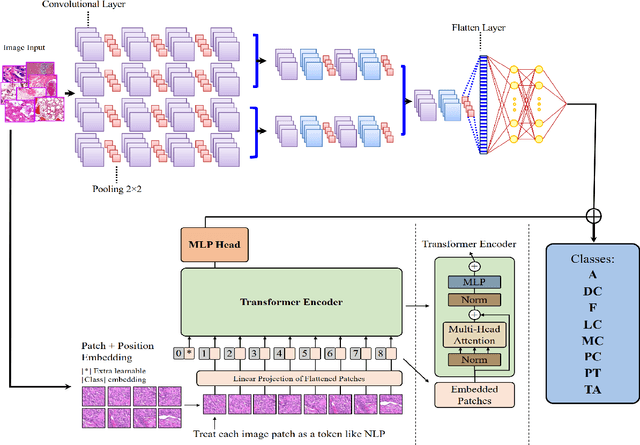
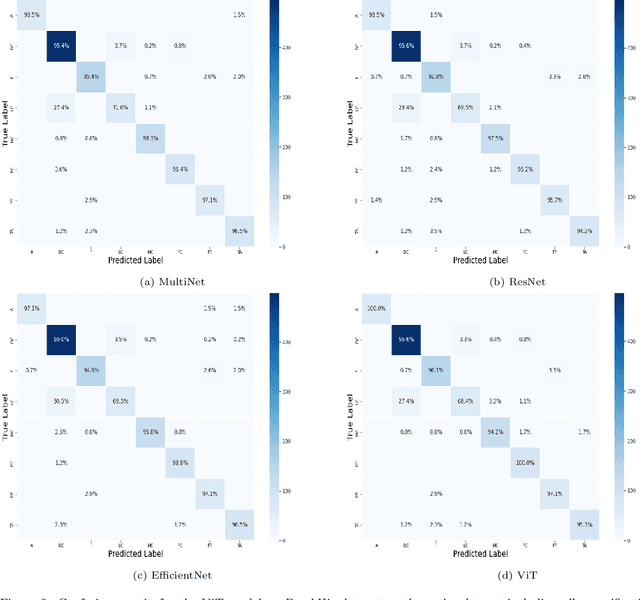
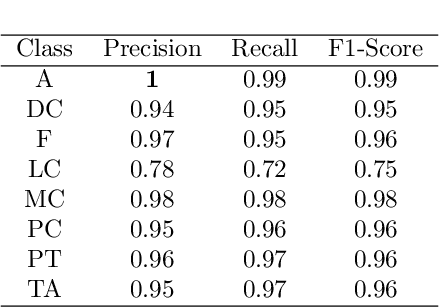
Abstract:Cancer is a leading cause of death in many countries. An early diagnosis of cancer based on biomedical imaging ensures effective treatment and a better prognosis. However, biomedical imaging presents challenges to both clinical institutions and researchers. Physiological anomalies are often characterized by slight abnormalities in individual cells or tissues, making them difficult to detect visually. Traditionally, anomalies are diagnosed by radiologists and pathologists with extensive training. This procedure, however, demands the participation of professionals and incurs a substantial cost. The cost makes large-scale biological image classification impractical. In this study, we provide unique deep neural network designs for multiclass classification of medical images, in particular cancer images. We incorporated transformers into a multiclass framework to take advantage of data-gathering capability and perform more accurate classifications. We evaluated models on publicly accessible datasets using various measures to ensure the reliability of the models. Extensive assessment metrics suggest this method can be used for a multitude of classification tasks.
Multiclass Semantic Segmentation to Identify Anatomical Sub-Regions of Brain and Measure Neuronal Health in Parkinson's Disease
Jan 07, 2023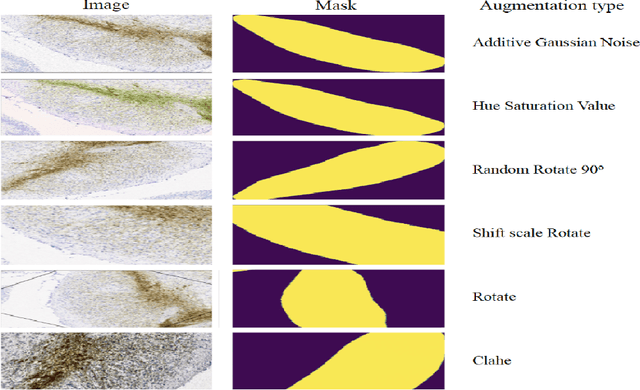
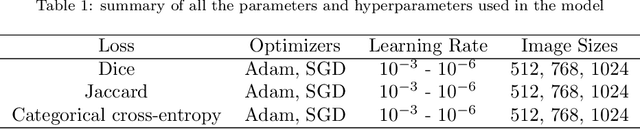
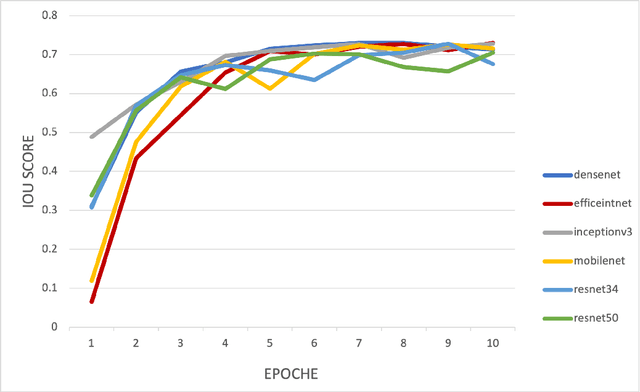

Abstract:Automated segmentation of anatomical sub-regions with high precision has become a necessity to enable the quantification and characterization of cells/ tissues in histology images. Currently, a machine learning model to analyze sub-anatomical regions of the brain to analyze 2D histological images is not available. The scientists rely on manually segmenting anatomical sub-regions of the brain which is extremely time-consuming and prone to labeler-dependent bias. One of the major challenges in accomplishing such a task is the lack of high-quality annotated images that can be used to train a generic artificial intelligence model. In this study, we employed a UNet-based architecture, compared model performance with various combinations of encoders, image sizes, and sample selection techniques. Additionally, to increase the sample set we resorted to data augmentation which provided data diversity and robust learning. In this study, we trained our best fit model on approximately one thousand annotated 2D brain images stained with Nissl/ Haematoxylin and Tyrosine Hydroxylase enzyme (TH, indicator of dopaminergic neuron viability). The dataset comprises of different animal studies enabling the model to be trained on different datasets. The model effectively is able to detect two sub-regions compacta (SNCD) and reticulata (SNr) in all the images. In spite of limited training data, our best model achieves a mean intersection over union (IOU) of 79% and a mean dice coefficient of 87%. In conclusion, the UNet-based model with EffiecientNet as an encoder outperforms all other encoders, resulting in a first of its kind robust model for multiclass segmentation of sub-brain regions in 2D images.
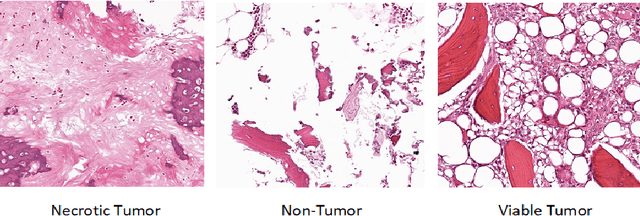
A Deep Learning Study on Osteosarcoma Detection from Histological Images
Nov 02, 2020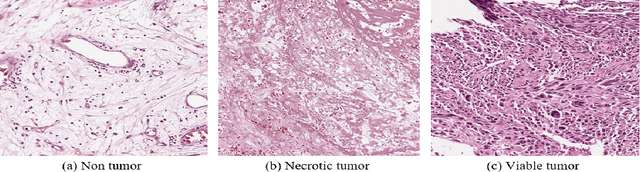
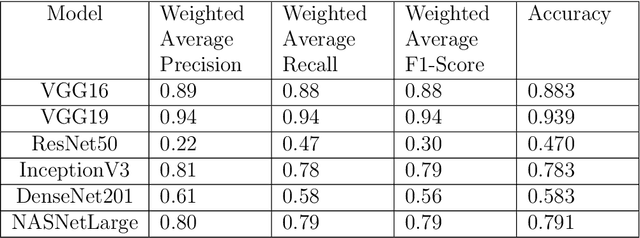
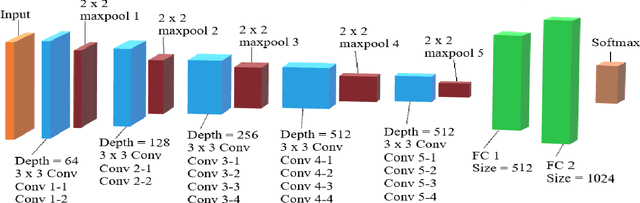
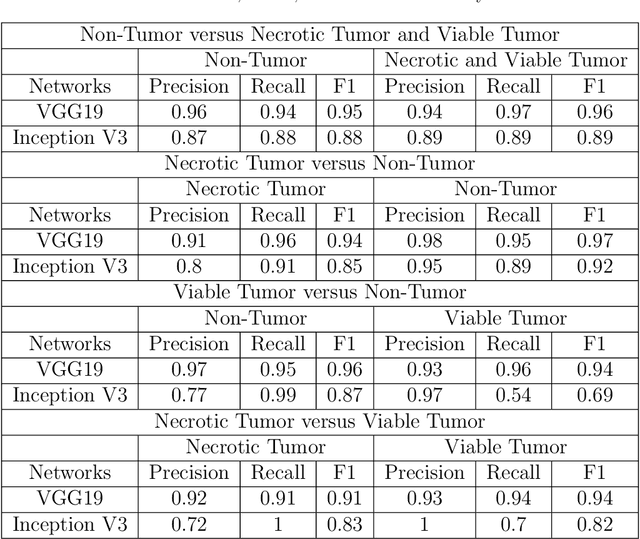
Abstract:In the U.S, 5-10\% of new pediatric cases of cancer are primary bone tumors. The most common type of primary malignant bone tumor is osteosarcoma. The intention of the present work is to improve the detection and diagnosis of osteosarcoma using computer-aided detection (CAD) and diagnosis (CADx). Such tools as convolutional neural networks (CNNs) can significantly decrease the surgeon's workload and make a better prognosis of patient conditions. CNNs need to be trained on a large amount of data in order to achieve a more trustworthy performance. In this study, transfer learning techniques, pre-trained CNNs, are adapted to a public dataset on osteosarcoma histological images to detect necrotic images from non-necrotic and healthy tissues. First, the dataset was preprocessed, and different classifications are applied. Then, Transfer learning models including VGG19 and Inception V3 are used and trained on Whole Slide Images (WSI) with no patches, to improve the accuracy of the outputs. Finally, the models are applied to different classification problems, including binary and multi-class classifiers. Experimental results show that the accuracy of the VGG19 has the highest, 96\%, performance amongst all binary classes and multiclass classification. Our fine-tuned model demonstrates state-of-the-art performance on detecting malignancy of Osteosarcoma based on histologic images.
C-Net: A Reliable Convolutional Neural Network for Biomedical Image Classification
Oct 30, 2020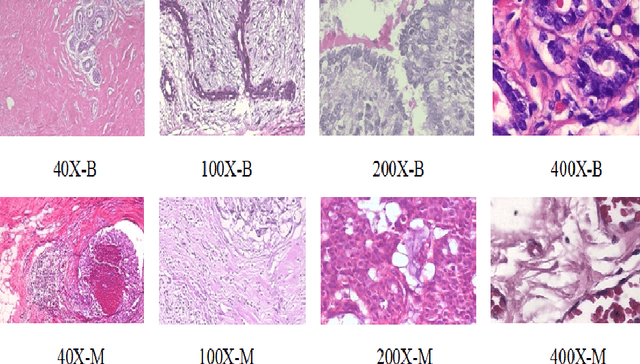
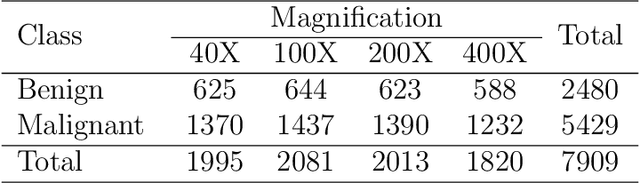
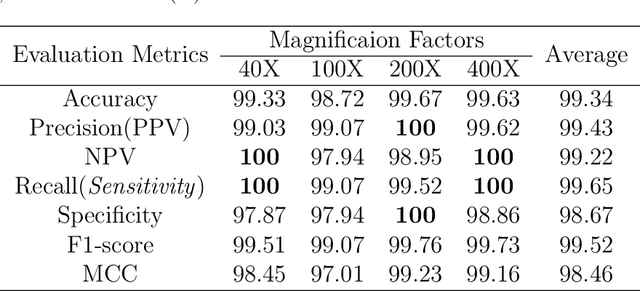
Abstract:Cancers are the leading cause of death in many developed countries. Early diagnosis plays a crucial role in having proper treatment for this debilitating disease. The automated classification of the type of cancer is a challenging task since pathologists must examine a huge number of histopathological images to detect infinitesimal abnormalities. In this study, we propose a novel convolutional neural network (CNN) architecture composed of a Concatenation of multiple Networks, called C-Net, to classify biomedical images. In contrast to conventional deep learning models in biomedical image classification, which utilize transfer learning to solve the problem, no prior knowledge is employed. The model incorporates multiple CNNs including Outer, Middle, and Inner. The first two parts of the architecture contain six networks that serve as feature extractors to feed into the Inner network to classify the images in terms of malignancy and benignancy. The C-Net is applied for histopathological image classification on two public datasets, including BreakHis and Osteosarcoma. To evaluate the performance, the model is tested using several evaluation metrics for its reliability. The C-Net model outperforms all other models on the individual metrics for both datasets and achieves zero misclassification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge