Valentina Pedoia
Foundations of a Knee Joint Digital Twin from qMRI Biomarkers for Osteoarthritis and Knee Replacement
Jan 26, 2025Abstract:This study forms the basis of a digital twin system of the knee joint, using advanced quantitative MRI (qMRI) and machine learning to advance precision health in osteoarthritis (OA) management and knee replacement (KR) prediction. We combined deep learning-based segmentation of knee joint structures with dimensionality reduction to create an embedded feature space of imaging biomarkers. Through cross-sectional cohort analysis and statistical modeling, we identified specific biomarkers, including variations in cartilage thickness and medial meniscus shape, that are significantly associated with OA incidence and KR outcomes. Integrating these findings into a comprehensive framework represents a considerable step toward personalized knee-joint digital twins, which could enhance therapeutic strategies and inform clinical decision-making in rheumatological care. This versatile and reliable infrastructure has the potential to be extended to broader clinical applications in precision health.
The object detection method aids in image reconstruction evaluation and clinical interpretation of meniscal abnormalities
Jul 16, 2024
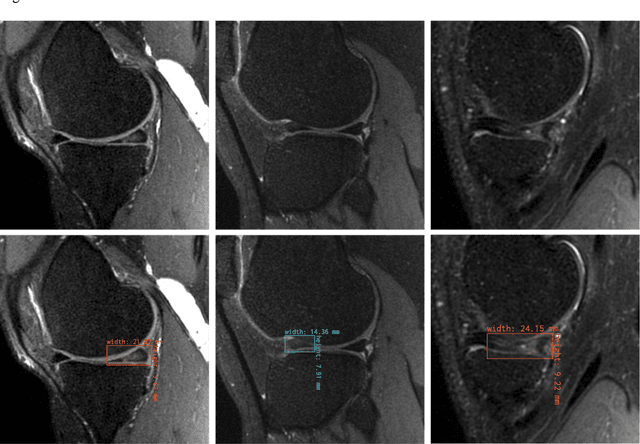
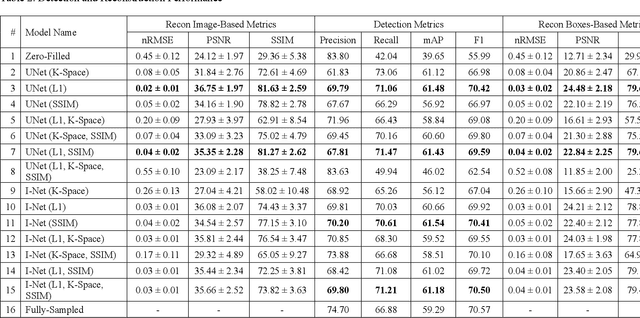
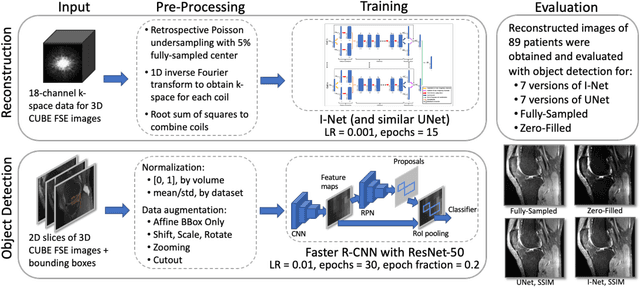
Abstract:This study investigates the relationship between deep learning (DL) image reconstruction quality and anomaly detection performance, and evaluates the efficacy of an artificial intelligence (AI) assistant in enhancing radiologists' interpretation of meniscal anomalies on reconstructed images. A retrospective study was conducted using an in-house reconstruction and anomaly detection pipeline to assess knee MR images from 896 patients. The original and 14 sets of DL-reconstructed images were evaluated using standard reconstruction and object detection metrics, alongside newly developed box-based reconstruction metrics. Two clinical radiologists reviewed a subset of 50 patients' images, both original and AI-assisted reconstructed, with subsequent assessment of their accuracy and performance characteristics. Results indicated that the structural similarity index (SSIM) showed a weaker correlation with anomaly detection metrics (mAP, r=0.64, p=0.01; F1 score, r=0.38, p=0.18), while box-based SSIM had a stronger association with detection performance (mAP, r=0.81, p<0.01; F1 score, r=0.65, p=0.01). Minor SSIM fluctuations did not affect detection outcomes, but significant changes reduced performance. Radiologists' AI-assisted evaluations demonstrated improved accuracy (86.0% without assistance vs. 88.3% with assistance, p<0.05) and interrater agreement (Cohen's kappa, 0.39 without assistance vs. 0.57 with assistance). An additional review led to the incorporation of 17 more lesions into the dataset. The proposed anomaly detection method shows promise in evaluating reconstruction algorithms for automated tasks and aiding radiologists in interpreting DL-reconstructed MR images.
Technical Note: Feasibility of translating 3.0T-trained Deep-Learning Segmentation Models Out-of-the-Box on Low-Field MRI 0.55T Knee-MRI of Healthy Controls
Oct 26, 2023Abstract:In the current study, our purpose is to evaluate the feasibility of applying deep learning (DL) enabled algorithms to quantify bilateral knee biomarkers in healthy controls scanned at 0.55T and compared with 3.0T. The current study assesses the performance of standard in-practice bone, and cartilage segmentation algorithms at 0.55T, both qualitatively and quantitatively, in terms of comparing segmentation performance, areas of improvement, and compartment-wise cartilage thickness values between 0.55T vs. 3.0T. Initial results demonstrate a usable to good technical feasibility of translating existing quantitative deep-learning-based image segmentation techniques, trained on 3.0T, out of 0.55T for knee MRI, in a multi-vendor acquisition environment. Especially in terms of segmenting cartilage compartments, the models perform almost equivalent to 3.0T in terms of Likert ranking. The 0.55T low-field sustainable and easy-to-install MRI, as demonstrated, thus, can be utilized for evaluating knee cartilage thickness and bone segmentations aided by established DL algorithms trained at higher-field strengths out-of-the-box initially. This could be utilized at the far-spread point-of-care locations with a lack of radiologists available to manually segment low-field images, at least till a decent base of low-field data pool is collated. With further fine-tuning with manual labeling of low-field data or utilizing synthesized higher SNR images from low-field images, OA biomarker quantification performance is potentially guaranteed to be further improved.
Leveraging wisdom of the crowds to improve consensus among radiologists by real time, blinded collaborations on a digital swarm platform
Jun 26, 2021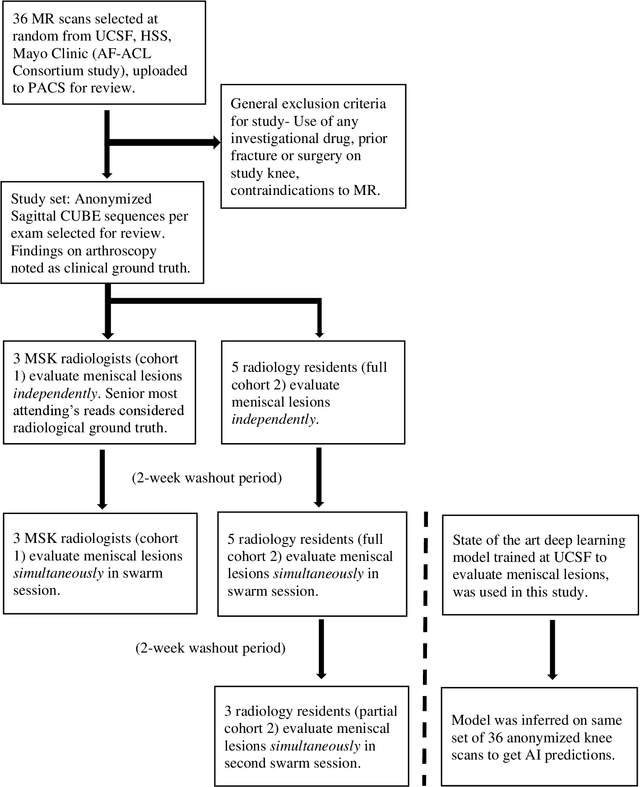
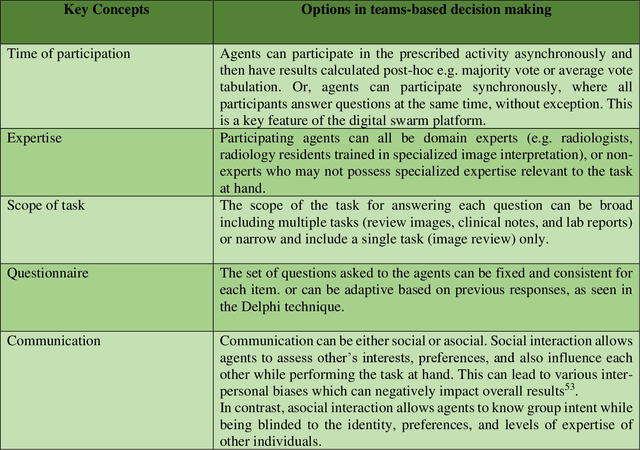
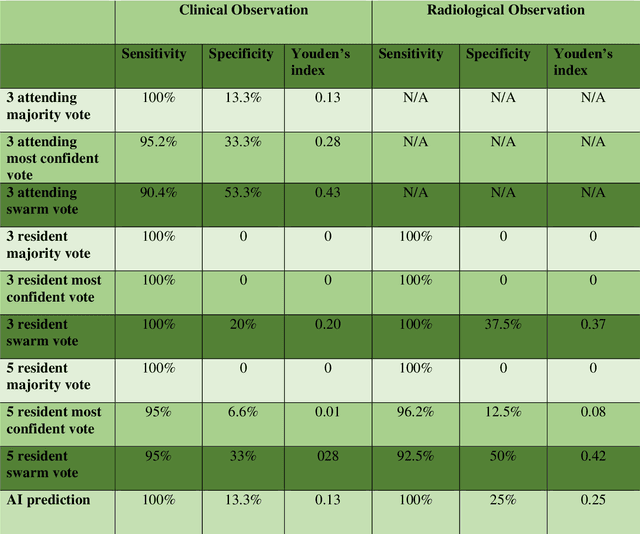
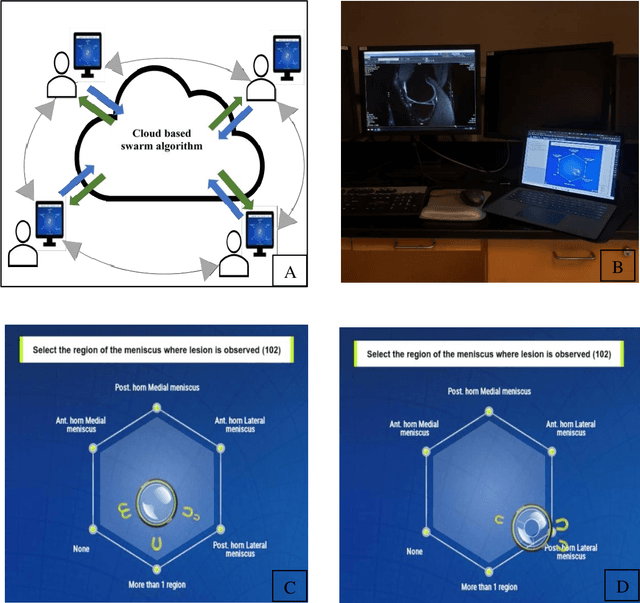
Abstract:Radiologists today play a key role in making diagnostic decisions and labeling images for training A.I. algorithms. Low inter-reader reliability (IRR) can be seen between experts when interpreting challenging cases. While teams-based decisions are known to outperform individual decisions, inter-personal biases often creep up in group interactions which limit non-dominant participants from expressing true opinions. To overcome the dual problems of low consensus and inter-personal bias, we explored a solution modeled on biological swarms of bees. Two separate cohorts; three radiologists and five radiology residents collaborated on a digital swarm platform in real time and in a blinded fashion, grading meniscal lesions on knee MR exams. These consensus votes were benchmarked against clinical (arthroscopy) and radiological (senior-most radiologist) observations. The IRR of the consensus votes was compared to the IRR of the majority and most confident votes of the two cohorts.The radiologist cohort saw an improvement of 23% in IRR of swarm votes over majority vote. Similar improvement of 23% in IRR in 3-resident swarm votes over majority vote, was observed. The 5-resident swarm had an even higher improvement of 32% in IRR over majority vote. Swarm consensus votes also improved specificity by up to 50%. The swarm consensus votes outperformed individual and majority vote decisions in both the radiologists and resident cohorts. The 5-resident swarm had higher IRR than 3-resident swarm indicating positive effect of increased swarm size. The attending and resident swarms also outperformed predictions from a state-of-the-art A.I. algorithm. Utilizing a digital swarm platform improved agreement and allows participants to express judgement free intent, resulting in superior clinical performance and robust A.I. training labels.
Adversarial Robust Training in MRI Reconstruction
Oct 30, 2020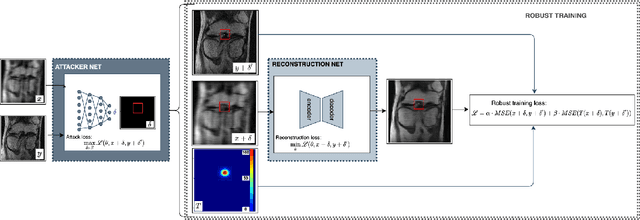
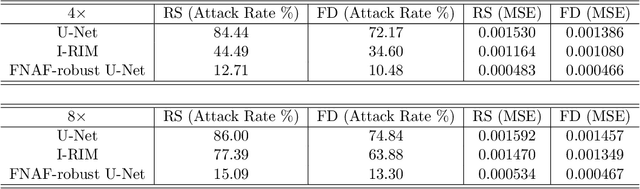
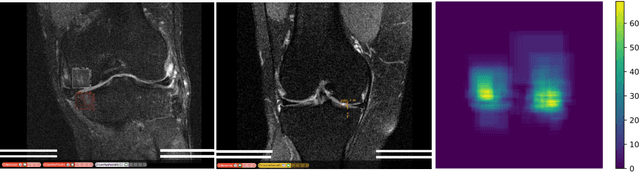
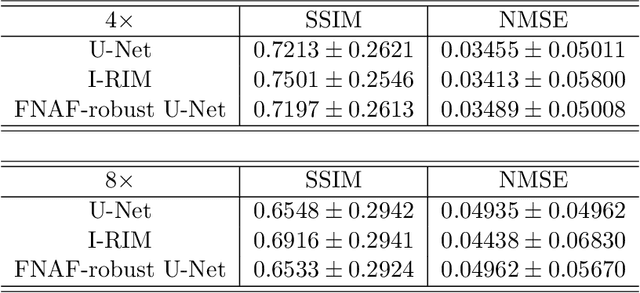
Abstract:Deep Learning has shown potential in accelerating Magnetic Resonance Image acquisition and reconstruction. Nevertheless, there is a dearth of tailored methods to guarantee that the reconstruction of small features is achieved with high fidelity. In this work, we employ adversarial attacks to generate small synthetic perturbations that when added to the input MRI, they are not reconstructed by a trained DL reconstruction network. Then, we use robust training to increase the network's sensitivity to small features and encourage their reconstruction. Next, we investigate the generalization of said approach to real world features. For this, a musculoskeletal radiologist annotated a set of cartilage and meniscal lesions from the knee Fast-MRI dataset, and a classification network was devised to assess the features reconstruction. Experimental results show that by introducing robust training to a reconstruction network, the rate (4.8\%) of false negative features in image reconstruction can be reduced. The results are encouraging and highlight the necessity for attention on this problem by the image reconstruction community, as a milestone for the introduction of DL reconstruction in clinical practice. To support further research, we make our annotation publicly available at https://github.com/fcaliva/fastMRI_BB_abnormalities_annotation.
The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset
May 26, 2020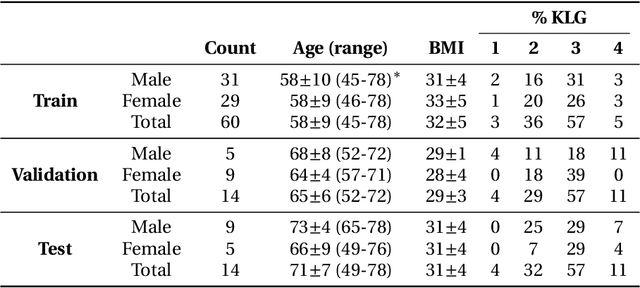
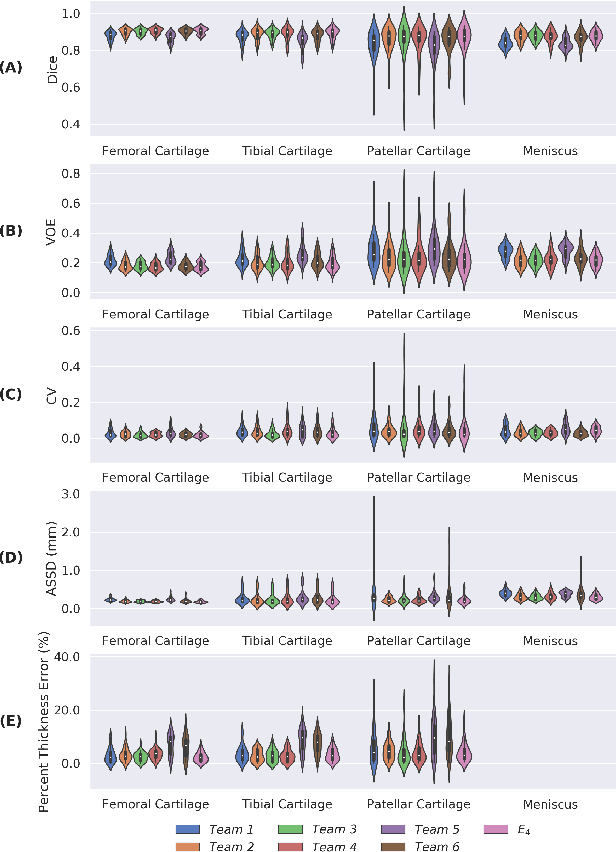
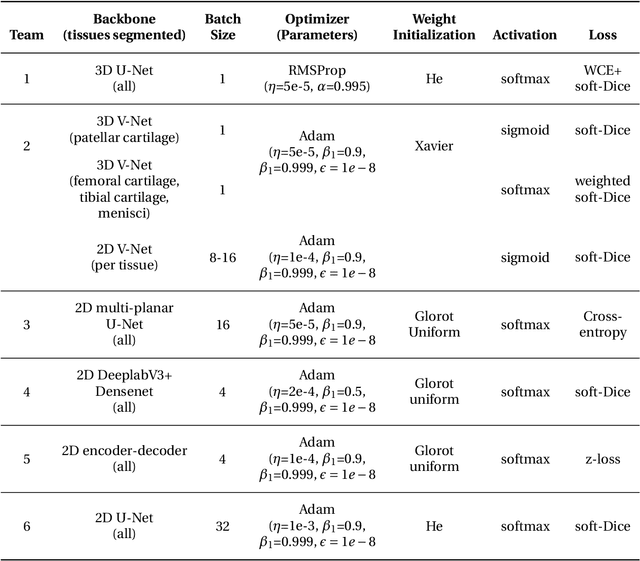
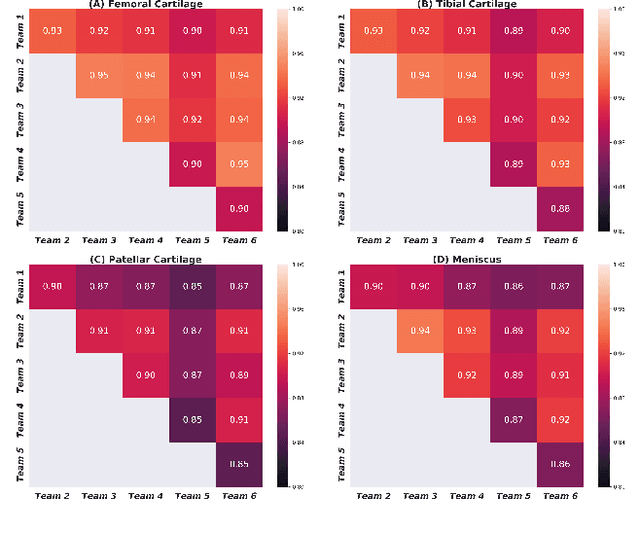
Abstract:Purpose: To organize a knee MRI segmentation challenge for characterizing the semantic and clinical efficacy of automatic segmentation methods relevant for monitoring osteoarthritis progression. Methods: A dataset partition consisting of 3D knee MRI from 88 subjects at two timepoints with ground-truth articular (femoral, tibial, patellar) cartilage and meniscus segmentations was standardized. Challenge submissions and a majority-vote ensemble were evaluated using Dice score, average symmetric surface distance, volumetric overlap error, and coefficient of variation on a hold-out test set. Similarities in network segmentations were evaluated using pairwise Dice correlations. Articular cartilage thickness was computed per-scan and longitudinally. Correlation between thickness error and segmentation metrics was measured using Pearson's coefficient. Two empirical upper bounds for ensemble performance were computed using combinations of model outputs that consolidated true positives and true negatives. Results: Six teams (T1-T6) submitted entries for the challenge. No significant differences were observed across all segmentation metrics for all tissues (p=1.0) among the four top-performing networks (T2, T3, T4, T6). Dice correlations between network pairs were high (>0.85). Per-scan thickness errors were negligible among T1-T4 (p=0.99) and longitudinal changes showed minimal bias (<0.03mm). Low correlations (<0.41) were observed between segmentation metrics and thickness error. The majority-vote ensemble was comparable to top performing networks (p=1.0). Empirical upper bound performances were similar for both combinations (p=1.0). Conclusion: Diverse networks learned to segment the knee similarly where high segmentation accuracy did not correlate to cartilage thickness accuracy. Voting ensembles did not outperform individual networks but may help regularize individual models.
Hierarchical Severity Staging of Anterior Cruciate Ligament Injuries using Deep Learning with MRI Images
Apr 13, 2020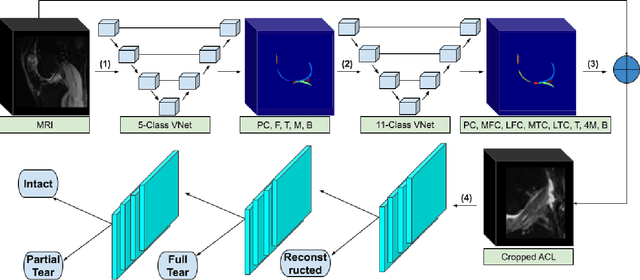
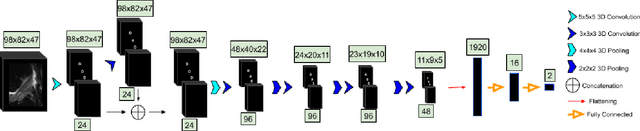
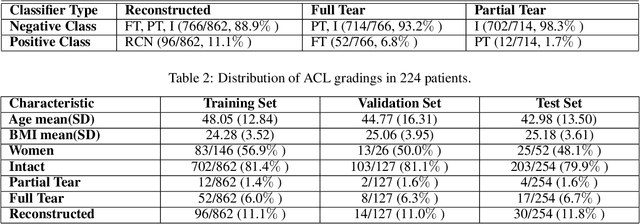
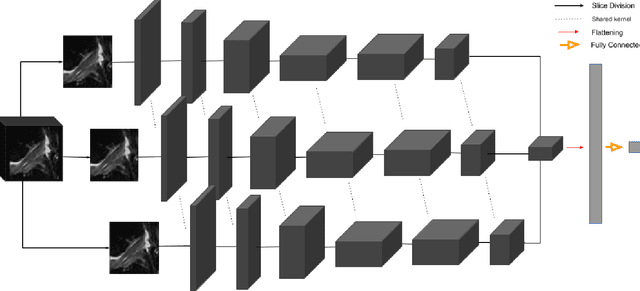
Abstract:Purpose: To evaluate the diagnostic utility of two convolutional neural networks (CNNs) for severity staging of anterior cruciate ligament (ACL) injuries. Materials and Methods: This retrospective analysis was conducted on 1243 knee MR images (1008 intact, 18 partially torn, 77 fully torn, and 140 reconstructed ACLs) from 224 patients (age 47 +/- 14 years, 54% women) acquired between 2011 and 2014. The radiologists used a modified scoring metric. To classify ACL injuries with deep learning, two types of CNNs were used, one with three-dimensional (3D) and the other with two-dimensional (2D) convolutional kernels. Performance metrics included sensitivity, specificity, weighted Cohen's kappa, and overall accuracy, followed by McNemar's test to compare the CNNs performance. Results: The overall accuracy and weighted Cohen's kappa reported for ACL injury classification were higher using the 2D CNN (accuracy: 92% (233/254) and kappa: 0.83) than the 3D CNN (accuracy: 89% (225/254) and kappa: 0.83) (P = .27). The 2D CNN and 3D CNN performed similarly in classifying intact ACLs (2D CNN: 93% (188/203) sensitivity and 90% (46/51) specificity; 3D CNN: 89% (180/203) sensitivity and 88% (45/51) specificity). Classification of full tears by both networks were also comparable (2D CNN: 82% (14/17) sensitivity and 94% (222/237) specificity; 3D CNN: 76% (13/17) sensitivity and 100% (236/237) specificity). The 2D CNN classified all reconstructed ACLs correctly. Conclusion: 2D and 3D CNNs applied to ACL lesion classification had high sensitivity and specificity, suggesting that these networks could be used to help grade ACL injuries by non-experts.
Deep learning predicts total knee replacement from magnetic resonance images
Feb 24, 2020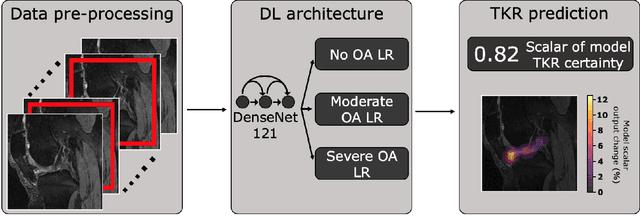
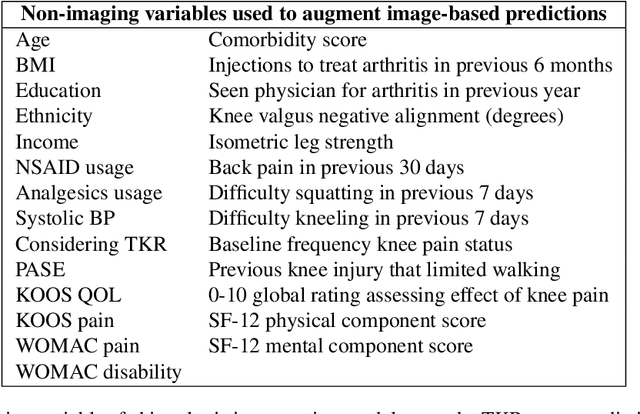

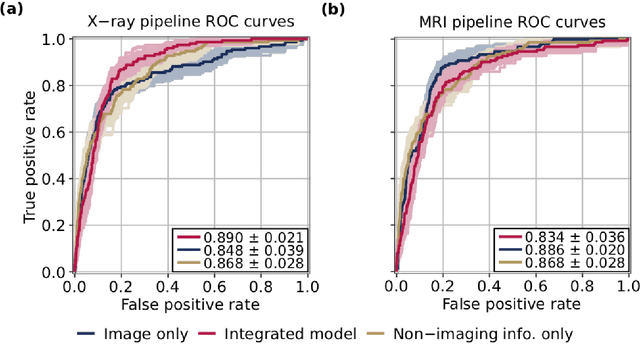
Abstract:Knee Osteoarthritis (OA) is a common musculoskeletal disorder in the United States. When diagnosed at early stages, lifestyle interventions such as exercise and weight loss can slow OA progression, but at later stages, only an invasive option is available: total knee replacement (TKR). Though a generally successful procedure, only 2/3 of patients who undergo the procedure report their knees feeling ''normal'' post-operation, and complications can arise that require revision. This necessitates a model to identify a population at higher risk of TKR, particularly at less advanced stages of OA, such that appropriate treatments can be implemented that slow OA progression and delay TKR. Here, we present a deep learning pipeline that leverages MRI images and clinical and demographic information to predict TKR with AUC $0.834 \pm 0.036$ (p < 0.05). Most notably, the pipeline predicts TKR with AUC $0.943 \pm 0.057$ (p < 0.05) for patients without OA. Furthermore, we develop occlusion maps for case-control pairs in test data and compare regions used by the model in both, thereby identifying TKR imaging biomarkers. As such, this work takes strides towards a pipeline with clinical utility, and the biomarkers identified further our understanding of OA progression and eventual TKR onset.
Automatic Hip Fracture Identification and Functional Subclassification with Deep Learning
Sep 10, 2019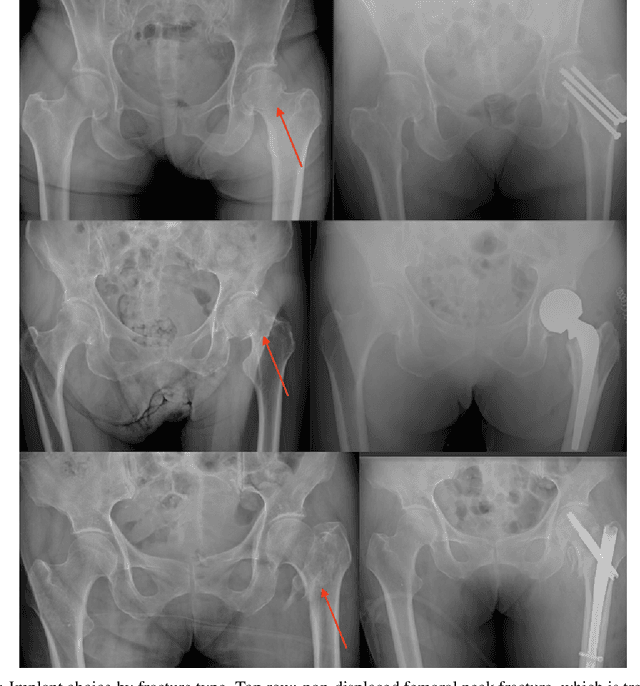
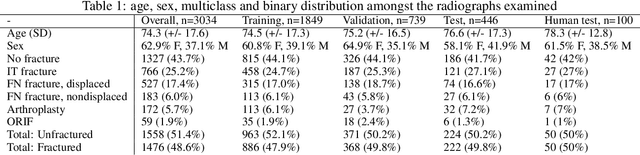
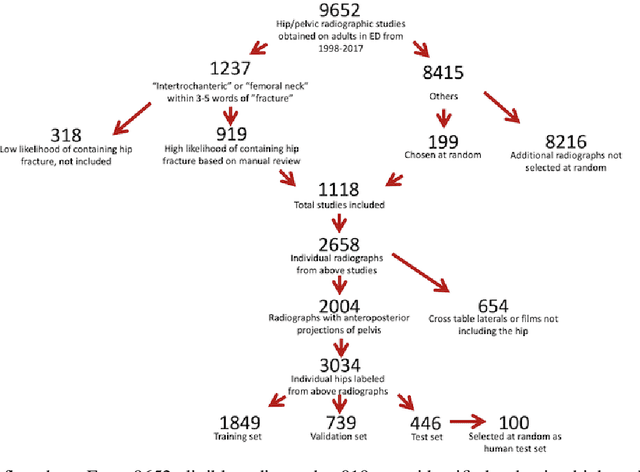
Abstract:Purpose: Hip fractures are a common cause of morbidity and mortality. Automatic identification and classification of hip fractures using deep learning may improve outcomes by reducing diagnostic errors and decreasing time to operation. Methods: Hip and pelvic radiographs from 1118 studies were reviewed and 3034 hips were labeled via bounding boxes and classified as normal, displaced femoral neck fracture, nondisplaced femoral neck fracture, intertrochanteric fracture, previous ORIF, or previous arthroplasty. A deep learning-based object detection model was trained to automate the placement of the bounding boxes. A Densely Connected Convolutional Neural Network (DenseNet) was trained on a subset of the bounding box images, and its performance evaluated on a held out test set and by comparison on a 100-image subset to two groups of human observers: fellowship-trained radiologists and orthopaedists, and senior residents in emergency medicine, radiology, and orthopaedics. Results: The binary accuracy for fracture of our model was 93.8% (95% CI, 91.3-95.8%), with sensitivity of 92.7% (95% CI, 88.7-95.6%), and specificity 95.0% (95% CI, 91.5-97.3%). Multiclass classification accuracy was 90.4% (95% CI, 87.4-92.9%). When compared to human observers, our model achieved at least expert-level classification under all conditions. Additionally, when the model was used as an aid, human performance improved, with aided resident performance approximating unaided fellowship-trained expert performance. Conclusions: Our deep learning model identified and classified hip fractures with at least expert-level accuracy, and when used as an aid improved human performance, with aided resident performance approximating that of unaided fellowship-trained attendings.
Adversarial Policy Gradient for Deep Learning Image Augmentation
Sep 09, 2019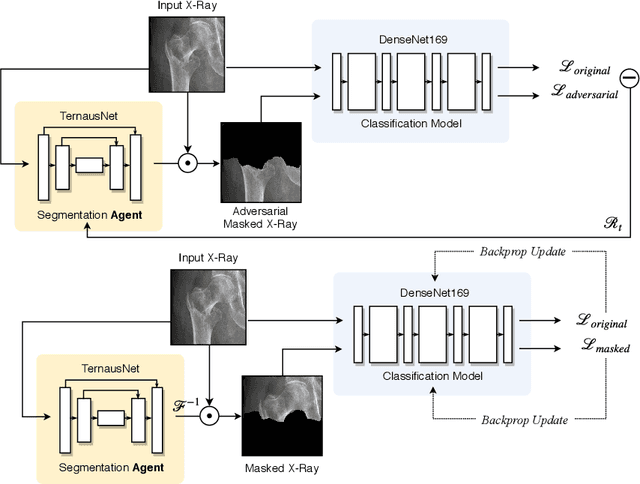
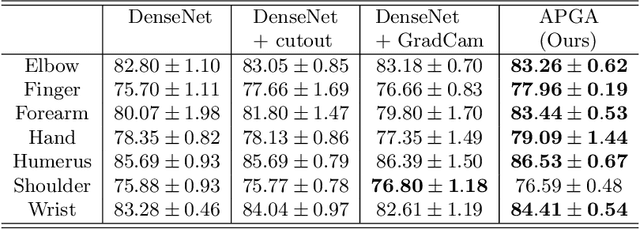
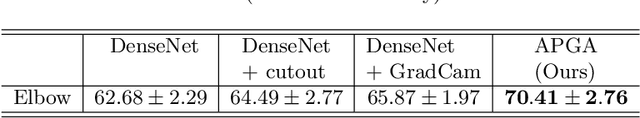
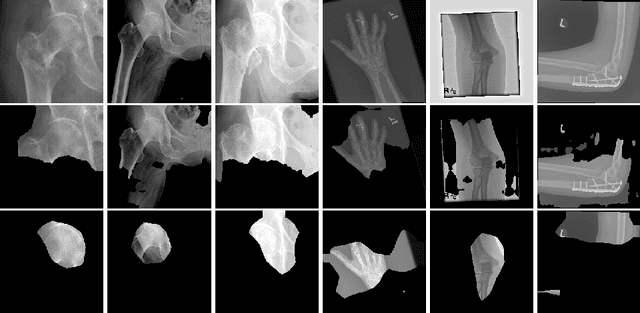
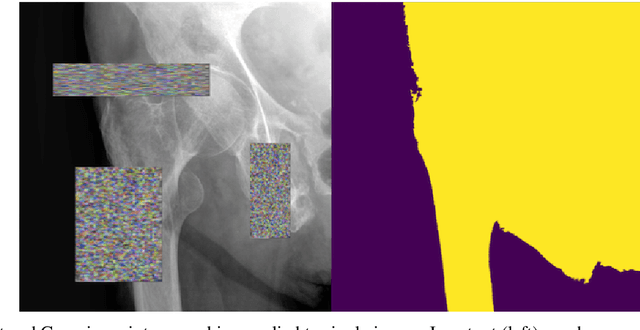
Abstract:The use of semantic segmentation for masking and cropping input images has proven to be a significant aid in medical imaging classification tasks by decreasing the noise and variance of the training dataset. However, implementing this approach with classical methods is challenging: the cost of obtaining a dense segmentation is high, and the precise input area that is most crucial to the classification task is difficult to determine a-priori. We propose a novel joint-training deep reinforcement learning framework for image augmentation. A segmentation network, weakly supervised with policy gradient optimization, acts as an agent, and outputs masks as actions given samples as states, with the goal of maximizing reward signals from the classification network. In this way, the segmentation network learns to mask unimportant imaging features. Our method, Adversarial Policy Gradient Augmentation (APGA), shows promising results on Stanford's MURA dataset and on a hip fracture classification task with an increase in global accuracy of up to 7.33% and improved performance over baseline methods in 9/10 tasks evaluated. We discuss the broad applicability of our joint training strategy to a variety of medical imaging tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge