Pan Su
LR-SQL: A Supervised Fine-Tuning Method for Text2SQL Tasks under Low-Resource Scenarios
Oct 15, 2024



Abstract:Large language models revolutionize Text2SQL through supervised fine-tuning, yet a crucial limitation is overlooked: the complexity of databases leads to an increased context length, consequently resulting in higher GPU memory demands for model fine-tuning. To address this issue, we propose LR-SQL. LR-SQL comprises two supervised fine-tuning models: the schema\_link model and the SQL\_generation model, with the schema\_link model serving as the focal point for streamlining the overall process. During the fine-tuning of the schema\_link model, LR-SQL breaks down the complete database into flexible combinations of tables with adjustable quantities, enabling the model to learn the relationships within the entire database from these dispersed slices. Furthermore, to enhance the model's ability to perceive the relationships among various discrete slices during inference, LR-SQL trains the model's Chain-of-Thought capability for this task. Experimental results demonstrate that LR-SQL can reduce the total GPU memory usage by 40\% compared to existing fine-tuning methods, while only losing 2\% of table prediction accuracy in schema\_link task. For the overall Text2SQL task, the Execution Accuracy decrease by 0.6\%.Our project is now available on https://github.com/hongWin/LR-SQL
Technical Note: Feasibility of translating 3.0T-trained Deep-Learning Segmentation Models Out-of-the-Box on Low-Field MRI 0.55T Knee-MRI of Healthy Controls
Oct 26, 2023Abstract:In the current study, our purpose is to evaluate the feasibility of applying deep learning (DL) enabled algorithms to quantify bilateral knee biomarkers in healthy controls scanned at 0.55T and compared with 3.0T. The current study assesses the performance of standard in-practice bone, and cartilage segmentation algorithms at 0.55T, both qualitatively and quantitatively, in terms of comparing segmentation performance, areas of improvement, and compartment-wise cartilage thickness values between 0.55T vs. 3.0T. Initial results demonstrate a usable to good technical feasibility of translating existing quantitative deep-learning-based image segmentation techniques, trained on 3.0T, out of 0.55T for knee MRI, in a multi-vendor acquisition environment. Especially in terms of segmenting cartilage compartments, the models perform almost equivalent to 3.0T in terms of Likert ranking. The 0.55T low-field sustainable and easy-to-install MRI, as demonstrated, thus, can be utilized for evaluating knee cartilage thickness and bone segmentations aided by established DL algorithms trained at higher-field strengths out-of-the-box initially. This could be utilized at the far-spread point-of-care locations with a lack of radiologists available to manually segment low-field images, at least till a decent base of low-field data pool is collated. With further fine-tuning with manual labeling of low-field data or utilizing synthesized higher SNR images from low-field images, OA biomarker quantification performance is potentially guaranteed to be further improved.
CS2-Net: Deep Learning Segmentation of Curvilinear Structures in Medical Imaging
Oct 19, 2020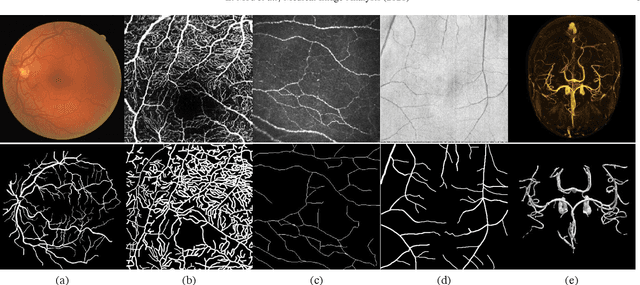
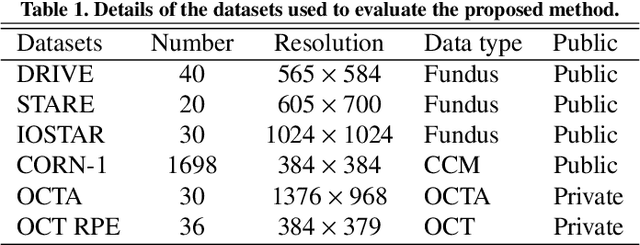
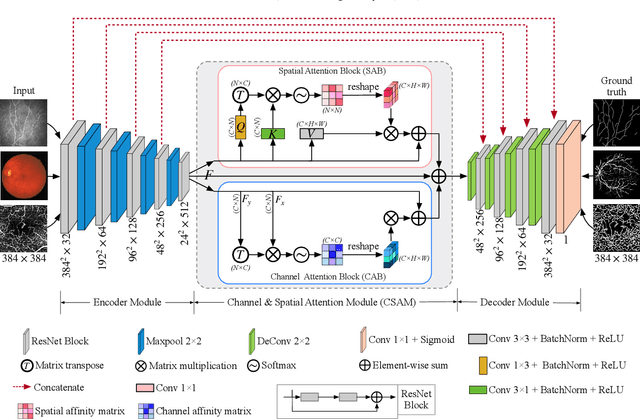
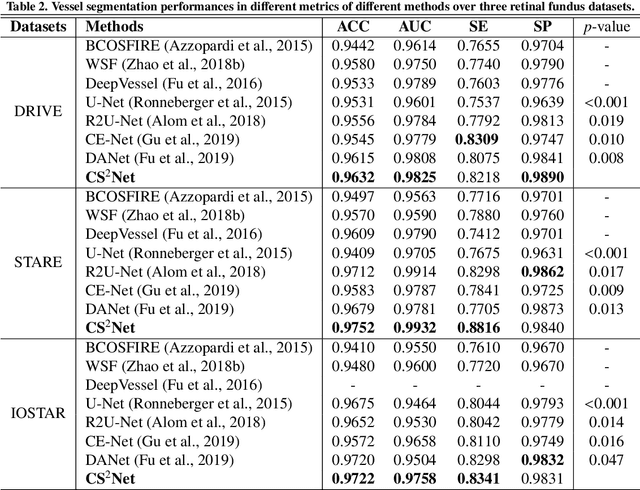
Abstract:Automated detection of curvilinear structures, e.g., blood vessels or nerve fibres, from medical and biomedical images is a crucial early step in automatic image interpretation associated to the management of many diseases. Precise measurement of the morphological changes of these curvilinear organ structures informs clinicians for understanding the mechanism, diagnosis, and treatment of e.g. cardiovascular, kidney, eye, lung, and neurological conditions. In this work, we propose a generic and unified convolution neural network for the segmentation of curvilinear structures and illustrate in several 2D/3D medical imaging modalities. We introduce a new curvilinear structure segmentation network (CS2-Net), which includes a self-attention mechanism in the encoder and decoder to learn rich hierarchical representations of curvilinear structures. Two types of attention modules - spatial attention and channel attention - are utilized to enhance the inter-class discrimination and intra-class responsiveness, to further integrate local features with their global dependencies and normalization, adaptively. Furthermore, to facilitate the segmentation of curvilinear structures in medical images, we employ a 1x3 and a 3x1 convolutional kernel to capture boundary features. ...
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge