Arjun D. Desai
Efficient Noise Calculation in Deep Learning-based MRI Reconstructions
May 04, 2025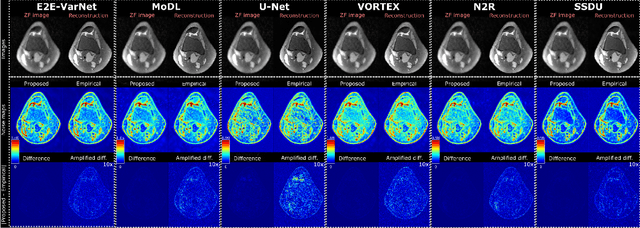
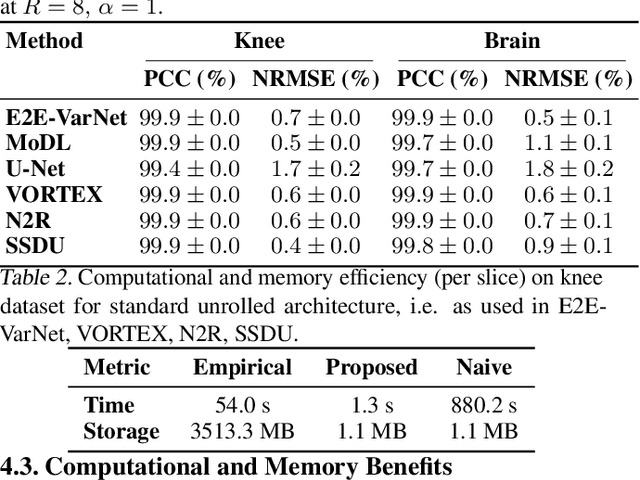
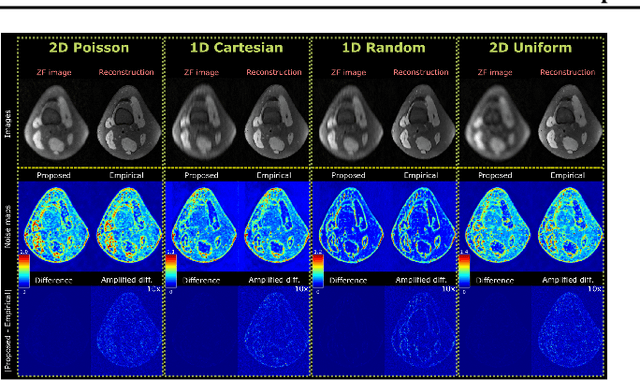
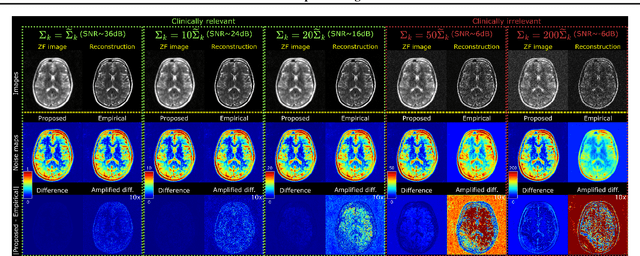
Abstract:Accelerated MRI reconstruction involves solving an ill-posed inverse problem where noise in acquired data propagates to the reconstructed images. Noise analyses are central to MRI reconstruction for providing an explicit measure of solution fidelity and for guiding the design and deployment of novel reconstruction methods. However, deep learning (DL)-based reconstruction methods have often overlooked noise propagation due to inherent analytical and computational challenges, despite its critical importance. This work proposes a theoretically grounded, memory-efficient technique to calculate voxel-wise variance for quantifying uncertainty due to acquisition noise in accelerated MRI reconstructions. Our approach approximates noise covariance using the DL network's Jacobian, which is intractable to calculate. To circumvent this, we derive an unbiased estimator for the diagonal of this covariance matrix (voxel-wise variance) and introduce a Jacobian sketching technique to efficiently implement it. We evaluate our method on knee and brain MRI datasets for both data- and physics-driven networks trained in supervised and unsupervised manners. Compared to empirical references obtained via Monte Carlo simulations, our technique achieves near-equivalent performance while reducing computational and memory demands by an order of magnitude or more. Furthermore, our method is robust across varying input noise levels, acceleration factors, and diverse undersampling schemes, highlighting its broad applicability. Our work reintroduces accurate and efficient noise analysis as a central tenet of reconstruction algorithms, holding promise to reshape how we evaluate and deploy DL-based MRI. Our code will be made publicly available upon acceptance.
Data-Limited Tissue Segmentation using Inpainting-Based Self-Supervised Learning
Oct 14, 2022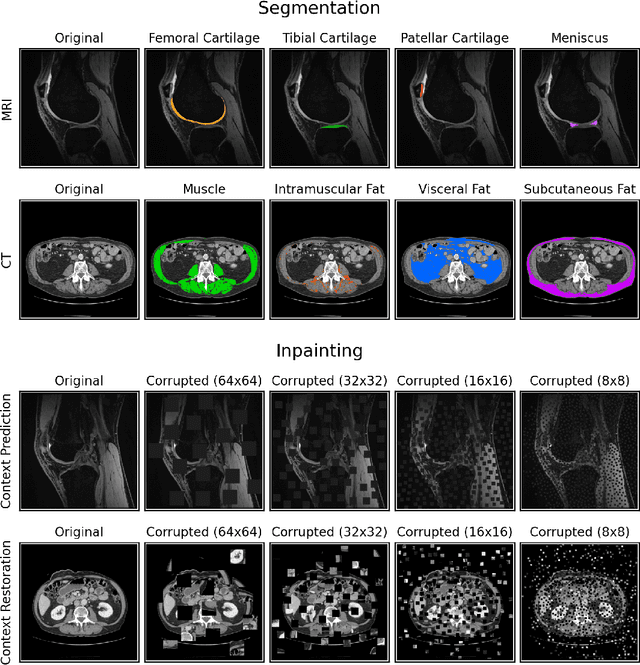
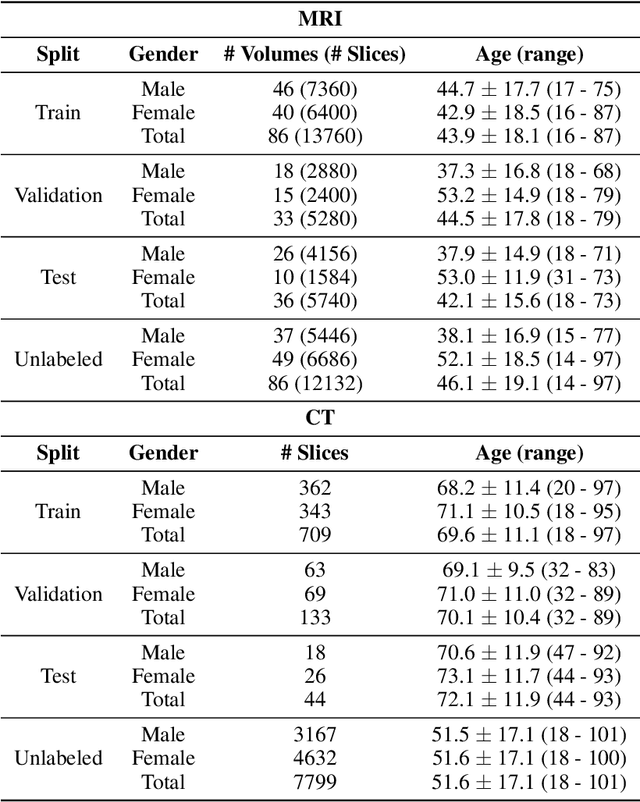
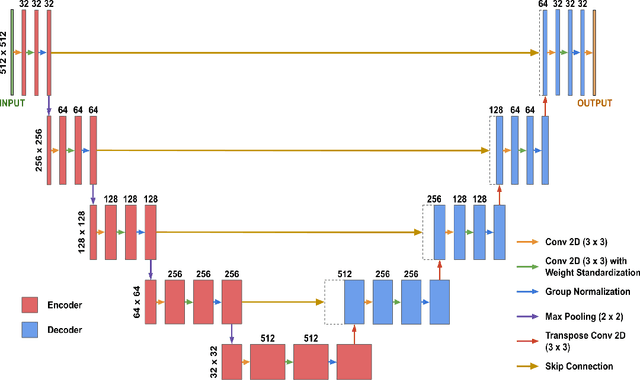
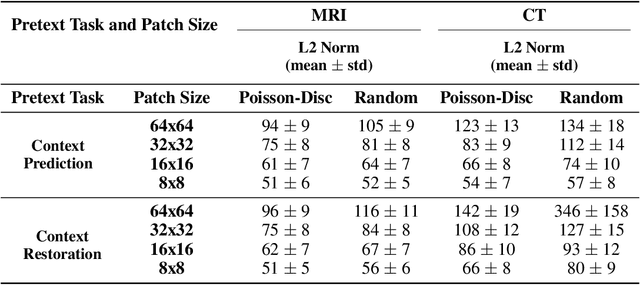
Abstract:Although supervised learning has enabled high performance for image segmentation, it requires a large amount of labeled training data, which can be difficult to obtain in the medical imaging field. Self-supervised learning (SSL) methods involving pretext tasks have shown promise in overcoming this requirement by first pretraining models using unlabeled data. In this work, we evaluate the efficacy of two SSL methods (inpainting-based pretext tasks of context prediction and context restoration) for CT and MRI image segmentation in label-limited scenarios, and investigate the effect of implementation design choices for SSL on downstream segmentation performance. We demonstrate that optimally trained and easy-to-implement inpainting-based SSL segmentation models can outperform classically supervised methods for MRI and CT tissue segmentation in label-limited scenarios, for both clinically-relevant metrics and the traditional Dice score.
Scale-Equivariant Unrolled Neural Networks for Data-Efficient Accelerated MRI Reconstruction
Apr 21, 2022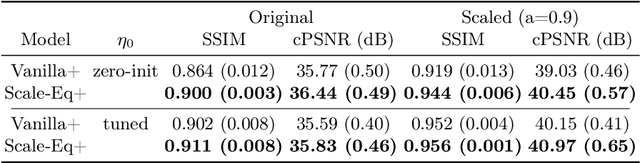
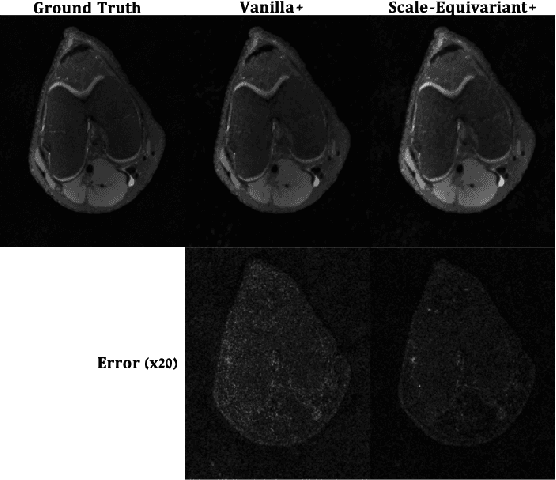
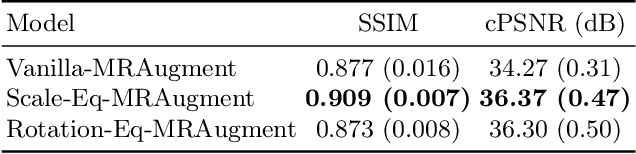
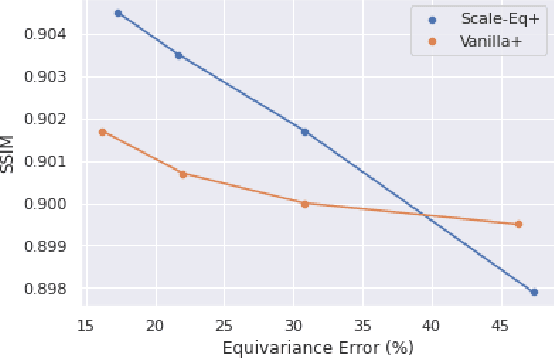
Abstract:Unrolled neural networks have enabled state-of-the-art reconstruction performance and fast inference times for the accelerated magnetic resonance imaging (MRI) reconstruction task. However, these approaches depend on fully-sampled scans as ground truth data which is either costly or not possible to acquire in many clinical medical imaging applications; hence, reducing dependence on data is desirable. In this work, we propose modeling the proximal operators of unrolled neural networks with scale-equivariant convolutional neural networks in order to improve the data-efficiency and robustness to drifts in scale of the images that might stem from the variability of patient anatomies or change in field-of-view across different MRI scanners. Our approach demonstrates strong improvements over the state-of-the-art unrolled neural networks under the same memory constraints both with and without data augmentations on both in-distribution and out-of-distribution scaled images without significantly increasing the train or inference time.
The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset
May 26, 2020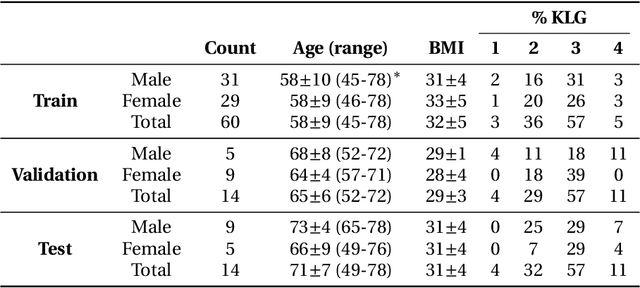
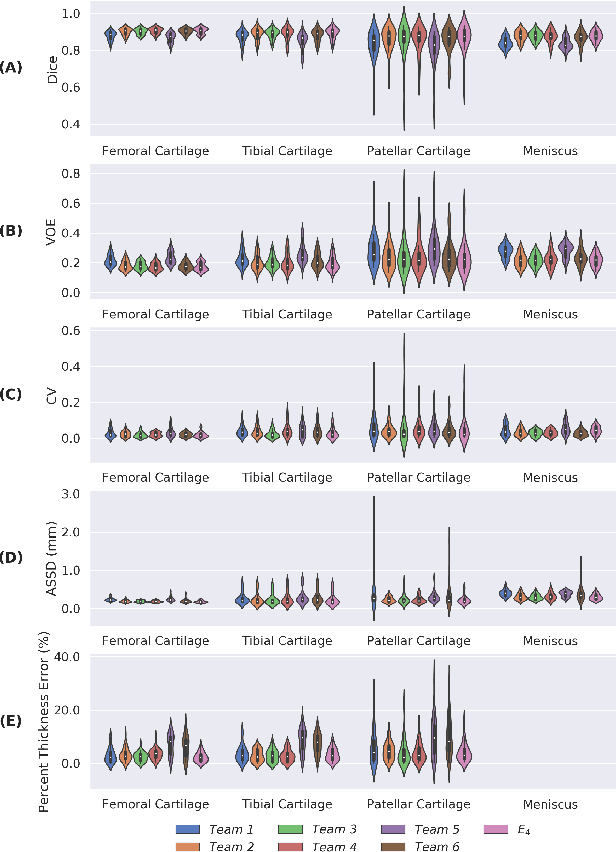
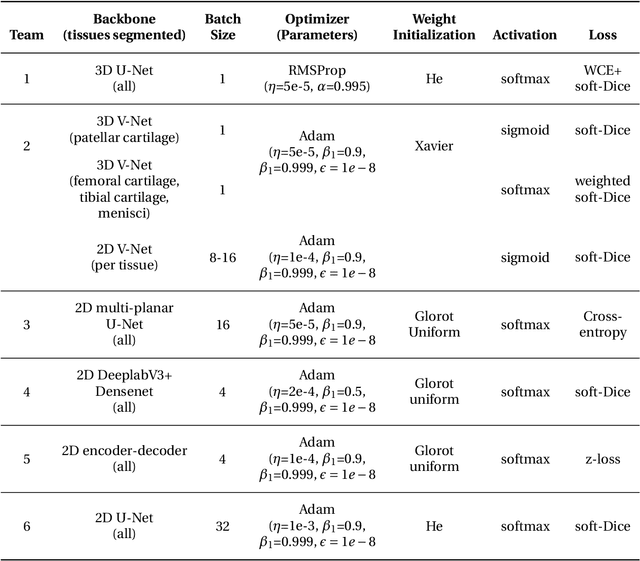
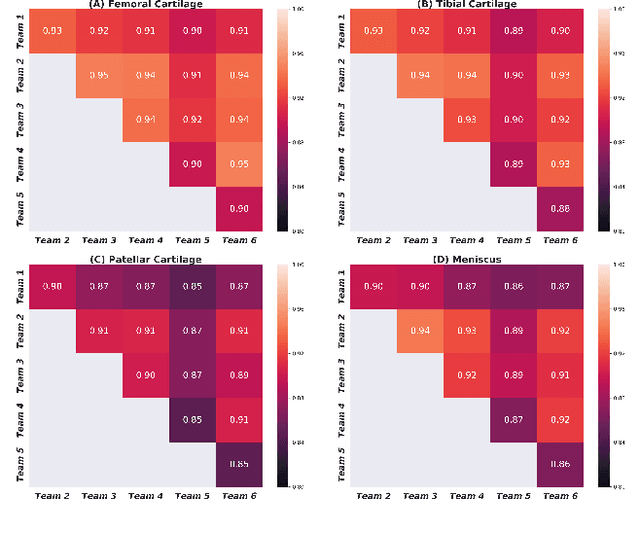
Abstract:Purpose: To organize a knee MRI segmentation challenge for characterizing the semantic and clinical efficacy of automatic segmentation methods relevant for monitoring osteoarthritis progression. Methods: A dataset partition consisting of 3D knee MRI from 88 subjects at two timepoints with ground-truth articular (femoral, tibial, patellar) cartilage and meniscus segmentations was standardized. Challenge submissions and a majority-vote ensemble were evaluated using Dice score, average symmetric surface distance, volumetric overlap error, and coefficient of variation on a hold-out test set. Similarities in network segmentations were evaluated using pairwise Dice correlations. Articular cartilage thickness was computed per-scan and longitudinally. Correlation between thickness error and segmentation metrics was measured using Pearson's coefficient. Two empirical upper bounds for ensemble performance were computed using combinations of model outputs that consolidated true positives and true negatives. Results: Six teams (T1-T6) submitted entries for the challenge. No significant differences were observed across all segmentation metrics for all tissues (p=1.0) among the four top-performing networks (T2, T3, T4, T6). Dice correlations between network pairs were high (>0.85). Per-scan thickness errors were negligible among T1-T4 (p=0.99) and longitudinal changes showed minimal bias (<0.03mm). Low correlations (<0.41) were observed between segmentation metrics and thickness error. The majority-vote ensemble was comparable to top performing networks (p=1.0). Empirical upper bound performances were similar for both combinations (p=1.0). Conclusion: Diverse networks learned to segment the knee similarly where high segmentation accuracy did not correlate to cartilage thickness accuracy. Voting ensembles did not outperform individual networks but may help regularize individual models.
Technical Considerations for Semantic Segmentation in MRI using Convolutional Neural Networks
Feb 05, 2019



Abstract:High-fidelity semantic segmentation of magnetic resonance volumes is critical for estimating tissue morphometry and relaxation parameters in both clinical and research applications. While manual segmentation is accepted as the gold-standard, recent advances in deep learning and convolutional neural networks (CNNs) have shown promise for efficient automatic segmentation of soft tissues. However, due to the stochastic nature of deep learning and the multitude of hyperparameters in training networks, predicting network behavior is challenging. In this paper, we quantify the impact of three factors associated with CNN segmentation performance: network architecture, training loss functions, and training data characteristics. We evaluate the impact of these variations on the segmentation of femoral cartilage and propose potential modifications to CNN architectures and training protocols to train these models with confidence.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge