Leon Lenchik
Comp2Comp: Open-Source Body Composition Assessment on Computed Tomography
Feb 13, 2023Abstract:Computed tomography (CT) is routinely used in clinical practice to evaluate a wide variety of medical conditions. While CT scans provide diagnoses, they also offer the ability to extract quantitative body composition metrics to analyze tissue volume and quality. Extracting quantitative body composition measures manually from CT scans is a cumbersome and time-consuming task. Proprietary software has been developed recently to automate this process, but the closed-source nature impedes widespread use. There is a growing need for fully automated body composition software that is more accessible and easier to use, especially for clinicians and researchers who are not experts in medical image processing. To this end, we have built Comp2Comp, an open-source Python package for rapid and automated body composition analysis of CT scans. This package offers models, post-processing heuristics, body composition metrics, automated batching, and polychromatic visualizations. Comp2Comp currently computes body composition measures for bone, skeletal muscle, visceral adipose tissue, and subcutaneous adipose tissue on CT scans of the abdomen. We have created two pipelines for this purpose. The first pipeline computes vertebral measures, as well as muscle and adipose tissue measures, at the T12 - L5 vertebral levels from abdominal CT scans. The second pipeline computes muscle and adipose tissue measures on user-specified 2D axial slices. In this guide, we discuss the architecture of the Comp2Comp pipelines, provide usage instructions, and report internal and external validation results to measure the quality of segmentations and body composition measures. Comp2Comp can be found at https://github.com/StanfordMIMI/Comp2Comp.
Data-Limited Tissue Segmentation using Inpainting-Based Self-Supervised Learning
Oct 14, 2022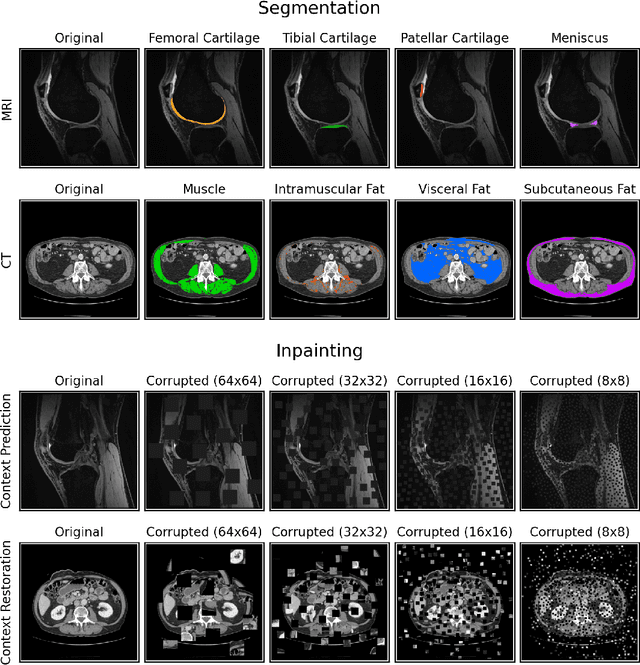
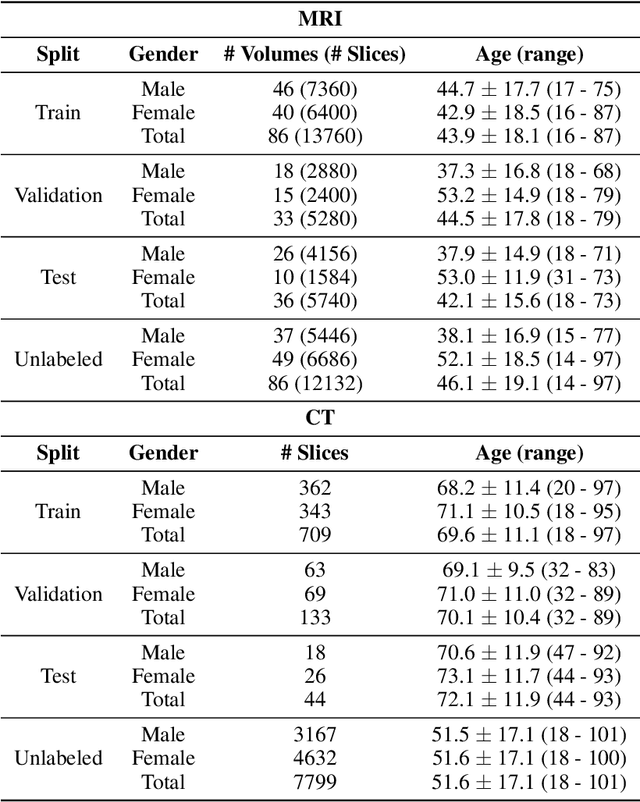
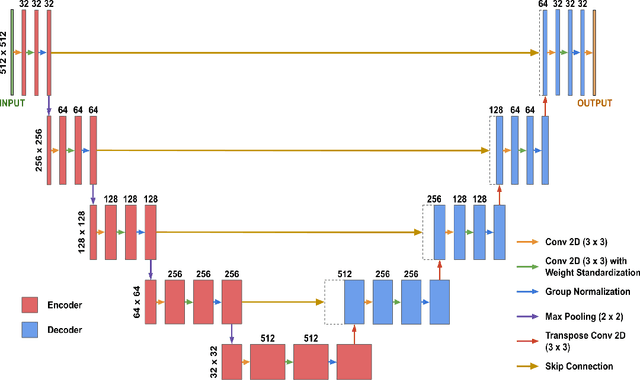
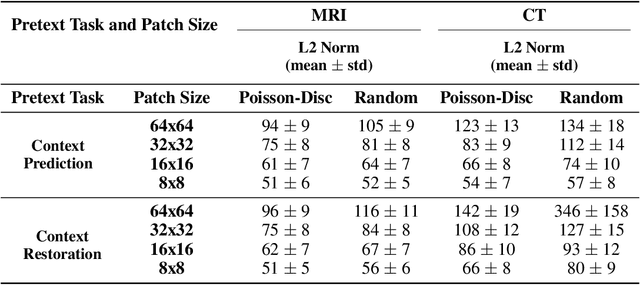
Abstract:Although supervised learning has enabled high performance for image segmentation, it requires a large amount of labeled training data, which can be difficult to obtain in the medical imaging field. Self-supervised learning (SSL) methods involving pretext tasks have shown promise in overcoming this requirement by first pretraining models using unlabeled data. In this work, we evaluate the efficacy of two SSL methods (inpainting-based pretext tasks of context prediction and context restoration) for CT and MRI image segmentation in label-limited scenarios, and investigate the effect of implementation design choices for SSL on downstream segmentation performance. We demonstrate that optimally trained and easy-to-implement inpainting-based SSL segmentation models can outperform classically supervised methods for MRI and CT tissue segmentation in label-limited scenarios, for both clinically-relevant metrics and the traditional Dice score.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge