Bhavik N. Patel
Comp2Comp: Open-Source Body Composition Assessment on Computed Tomography
Feb 13, 2023Abstract:Computed tomography (CT) is routinely used in clinical practice to evaluate a wide variety of medical conditions. While CT scans provide diagnoses, they also offer the ability to extract quantitative body composition metrics to analyze tissue volume and quality. Extracting quantitative body composition measures manually from CT scans is a cumbersome and time-consuming task. Proprietary software has been developed recently to automate this process, but the closed-source nature impedes widespread use. There is a growing need for fully automated body composition software that is more accessible and easier to use, especially for clinicians and researchers who are not experts in medical image processing. To this end, we have built Comp2Comp, an open-source Python package for rapid and automated body composition analysis of CT scans. This package offers models, post-processing heuristics, body composition metrics, automated batching, and polychromatic visualizations. Comp2Comp currently computes body composition measures for bone, skeletal muscle, visceral adipose tissue, and subcutaneous adipose tissue on CT scans of the abdomen. We have created two pipelines for this purpose. The first pipeline computes vertebral measures, as well as muscle and adipose tissue measures, at the T12 - L5 vertebral levels from abdominal CT scans. The second pipeline computes muscle and adipose tissue measures on user-specified 2D axial slices. In this guide, we discuss the architecture of the Comp2Comp pipelines, provide usage instructions, and report internal and external validation results to measure the quality of segmentations and body composition measures. Comp2Comp can be found at https://github.com/StanfordMIMI/Comp2Comp.
Multimodal spatiotemporal graph neural networks for improved prediction of 30-day all-cause hospital readmission
Apr 14, 2022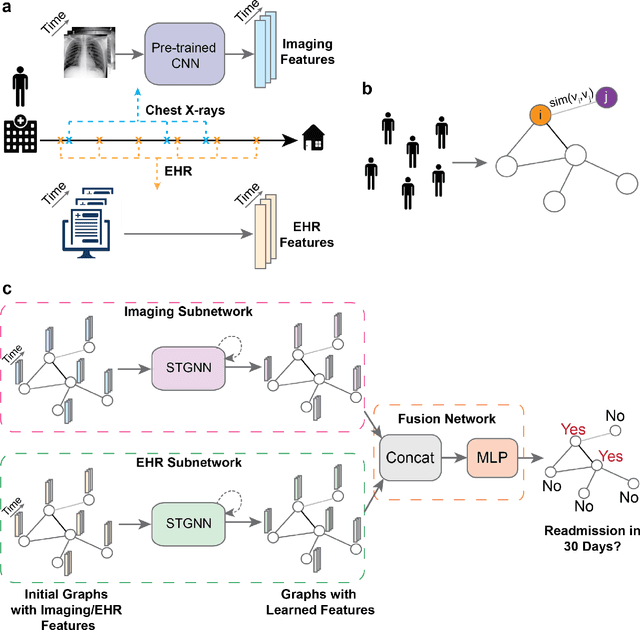
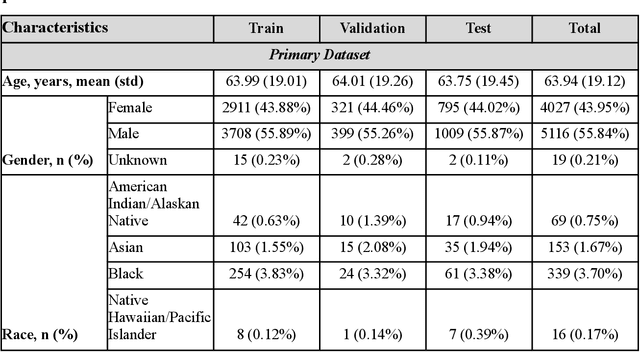
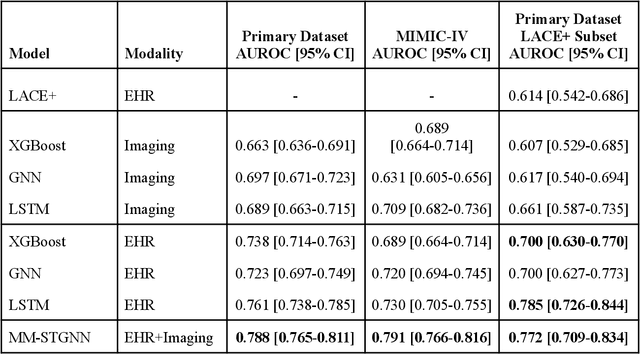
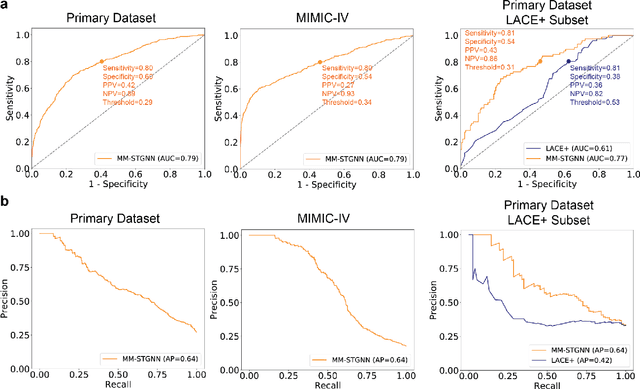
Abstract:Measures to predict 30-day readmission are considered an important quality factor for hospitals as accurate predictions can reduce the overall cost of care by identifying high risk patients before they are discharged. While recent deep learning-based studies have shown promising empirical results on readmission prediction, several limitations exist that may hinder widespread clinical utility, such as (a) only patients with certain conditions are considered, (b) existing approaches do not leverage data temporality, (c) individual admissions are assumed independent of each other, which is unrealistic, (d) prior studies are usually limited to single source of data and single center data. To address these limitations, we propose a multimodal, modality-agnostic spatiotemporal graph neural network (MM-STGNN) for prediction of 30-day all-cause hospital readmission that fuses multimodal in-patient longitudinal data. By training and evaluating our methods using longitudinal chest radiographs and electronic health records from two independent centers, we demonstrate that MM-STGNN achieves AUROC of 0.79 on both primary and external datasets. Furthermore, MM-STGNN significantly outperforms the current clinical reference standard, LACE+ score (AUROC=0.61), on the primary dataset. For subset populations of patients with heart and vascular disease, our model also outperforms baselines on predicting 30-day readmission (e.g., 3.7 point improvement in AUROC in patients with heart disease). Lastly, qualitative model interpretability analysis indicates that while patients' primary diagnoses were not explicitly used to train the model, node features crucial for model prediction directly reflect patients' primary diagnoses. Importantly, our MM-STGNN is agnostic to node feature modalities and could be utilized to integrate multimodal data for triaging patients in various downstream resource allocation tasks.
Assessing Robustness to Noise: Low-Cost Head CT Triage
Mar 29, 2020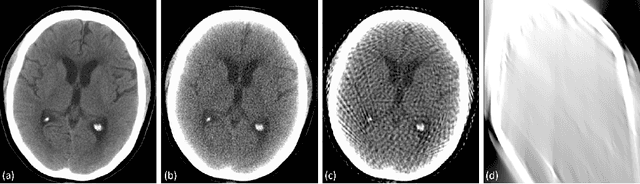
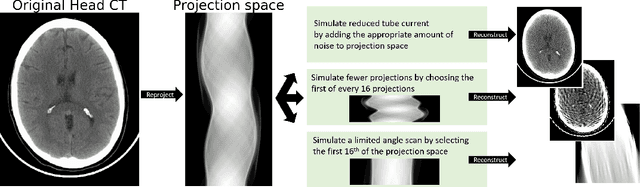
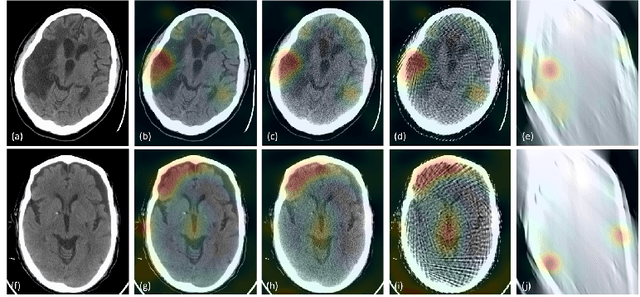
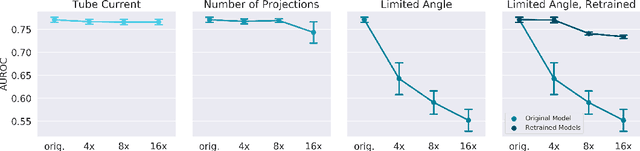
Abstract:Automated medical image classification with convolutional neural networks (CNNs) has great potential to impact healthcare, particularly in resource-constrained healthcare systems where fewer trained radiologists are available. However, little is known about how well a trained CNN can perform on images with the increased noise levels, different acquisition protocols, or additional artifacts that may arise when using low-cost scanners, which can be underrepresented in datasets collected from well-funded hospitals. In this work, we investigate how a model trained to triage head computed tomography (CT) scans performs on images acquired with reduced x-ray tube current, fewer projections per gantry rotation, and limited angle scans. These changes can reduce the cost of the scanner and demands on electrical power but come at the expense of increased image noise and artifacts. We first develop a model to triage head CTs and report an area under the receiver operating characteristic curve (AUROC) of 0.77. We then show that the trained model is robust to reduced tube current and fewer projections, with the AUROC dropping only 0.65% for images acquired with a 16x reduction in tube current and 0.22% for images acquired with 8x fewer projections. Finally, for significantly degraded images acquired by a limited angle scan, we show that a model trained specifically to classify such images can overcome the technological limitations to reconstruction and maintain an AUROC within 0.09% of the original model.
CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison
Jan 21, 2019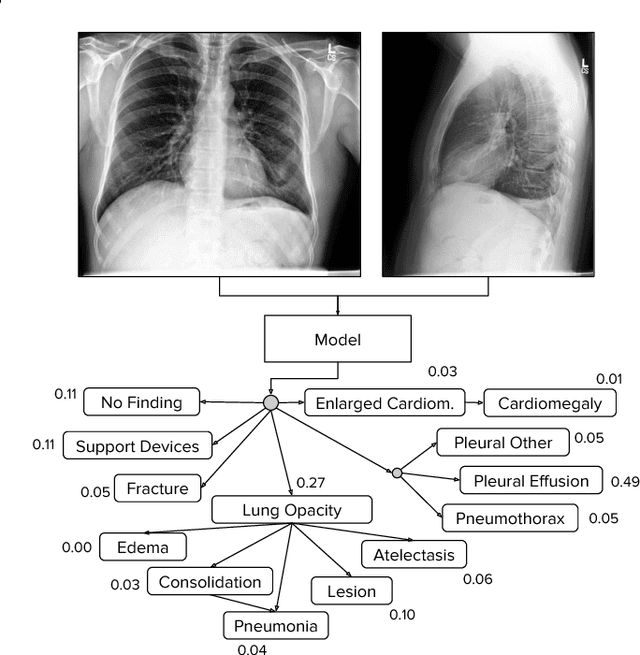
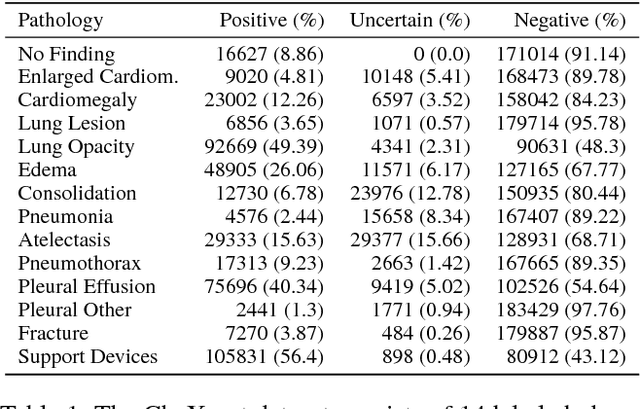
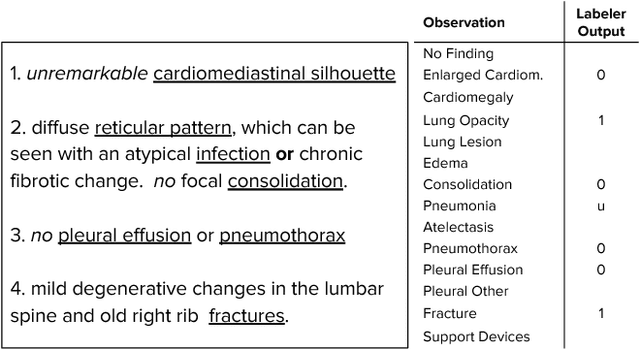
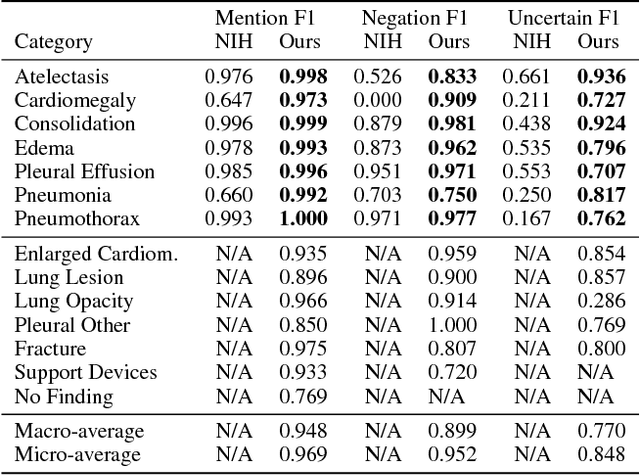
Abstract:Large, labeled datasets have driven deep learning methods to achieve expert-level performance on a variety of medical imaging tasks. We present CheXpert, a large dataset that contains 224,316 chest radiographs of 65,240 patients. We design a labeler to automatically detect the presence of 14 observations in radiology reports, capturing uncertainties inherent in radiograph interpretation. We investigate different approaches to using the uncertainty labels for training convolutional neural networks that output the probability of these observations given the available frontal and lateral radiographs. On a validation set of 200 chest radiographic studies which were manually annotated by 3 board-certified radiologists, we find that different uncertainty approaches are useful for different pathologies. We then evaluate our best model on a test set composed of 500 chest radiographic studies annotated by a consensus of 5 board-certified radiologists, and compare the performance of our model to that of 3 additional radiologists in the detection of 5 selected pathologies. On Cardiomegaly, Edema, and Pleural Effusion, the model ROC and PR curves lie above all 3 radiologist operating points. We release the dataset to the public as a standard benchmark to evaluate performance of chest radiograph interpretation models. The dataset is freely available at https://stanfordmlgroup.github.io/competitions/chexpert .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge