Jared Dunnmon
Advancing the Scientific Method with Large Language Models: From Hypothesis to Discovery
May 22, 2025Abstract:With recent Nobel Prizes recognising AI contributions to science, Large Language Models (LLMs) are transforming scientific research by enhancing productivity and reshaping the scientific method. LLMs are now involved in experimental design, data analysis, and workflows, particularly in chemistry and biology. However, challenges such as hallucinations and reliability persist. In this contribution, we review how Large Language Models (LLMs) are redefining the scientific method and explore their potential applications across different stages of the scientific cycle, from hypothesis testing to discovery. We conclude that, for LLMs to serve as relevant and effective creative engines and productivity enhancers, their deep integration into all steps of the scientific process should be pursued in collaboration and alignment with human scientific goals, with clear evaluation metrics. The transition to AI-driven science raises ethical questions about creativity, oversight, and responsibility. With careful guidance, LLMs could evolve into creative engines, driving transformative breakthroughs across scientific disciplines responsibly and effectively. However, the scientific community must also decide how much it leaves to LLMs to drive science, even when associations with 'reasoning', mostly currently undeserved, are made in exchange for the potential to explore hypothesis and solution regions that might otherwise remain unexplored by human exploration alone.
* 45 pages
xView3-SAR: Detecting Dark Fishing Activity Using Synthetic Aperture Imagery
Jun 02, 2022
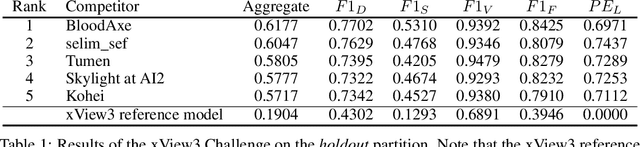

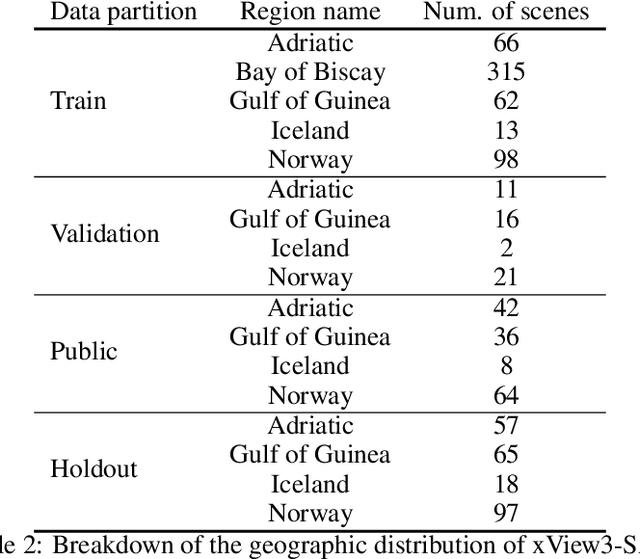
Abstract:Unsustainable fishing practices worldwide pose a major threat to marine resources and ecosystems. Identifying vessels that evade monitoring systems -- known as "dark vessels" -- is key to managing and securing the health of marine environments. With the rise of satellite-based synthetic aperture radar (SAR) imaging and modern machine learning (ML), it is now possible to automate detection of dark vessels day or night, under all-weather conditions. SAR images, however, require domain-specific treatment and is not widely accessible to the ML community. Moreover, the objects (vessels) are small and sparse, challenging traditional computer vision approaches. We present the largest labeled dataset for training ML models to detect and characterize vessels from SAR. xView3-SAR consists of nearly 1,000 analysis-ready SAR images from the Sentinel-1 mission that are, on average, 29,400-by-24,400 pixels each. The images are annotated using a combination of automated and manual analysis. Co-located bathymetry and wind state rasters accompany every SAR image. We provide an overview of the results from the xView3 Computer Vision Challenge, an international competition using xView3-SAR for ship detection and characterization at large scale. We release the data (https://iuu.xview.us/) and code (https://github.com/DIUx-xView) to support ongoing development and evaluation of ML approaches for this important application.
Multimodal spatiotemporal graph neural networks for improved prediction of 30-day all-cause hospital readmission
Apr 14, 2022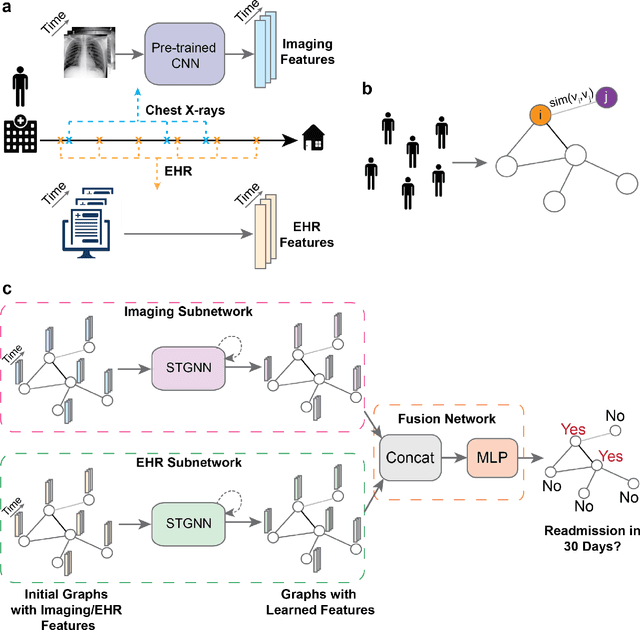
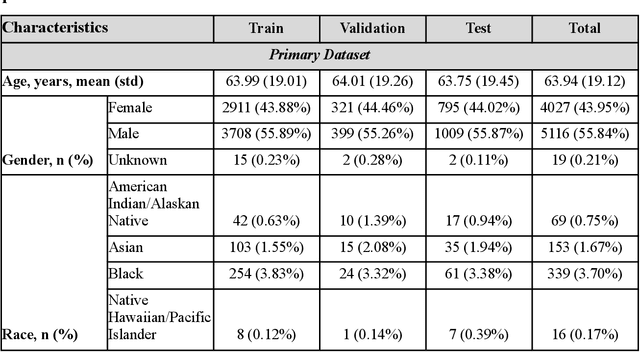
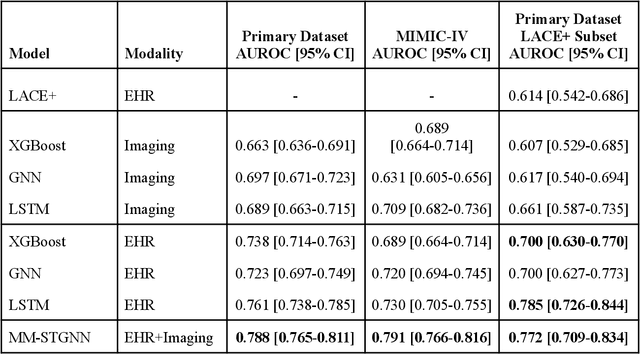
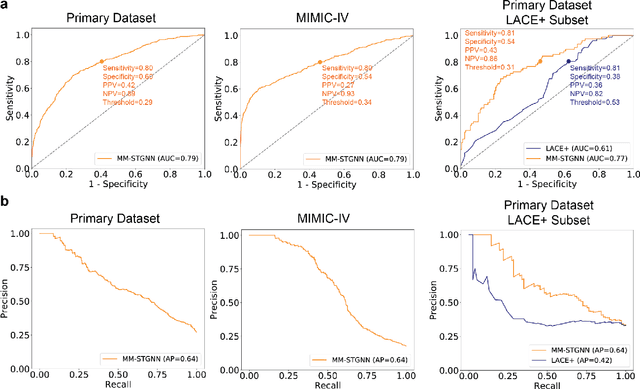
Abstract:Measures to predict 30-day readmission are considered an important quality factor for hospitals as accurate predictions can reduce the overall cost of care by identifying high risk patients before they are discharged. While recent deep learning-based studies have shown promising empirical results on readmission prediction, several limitations exist that may hinder widespread clinical utility, such as (a) only patients with certain conditions are considered, (b) existing approaches do not leverage data temporality, (c) individual admissions are assumed independent of each other, which is unrealistic, (d) prior studies are usually limited to single source of data and single center data. To address these limitations, we propose a multimodal, modality-agnostic spatiotemporal graph neural network (MM-STGNN) for prediction of 30-day all-cause hospital readmission that fuses multimodal in-patient longitudinal data. By training and evaluating our methods using longitudinal chest radiographs and electronic health records from two independent centers, we demonstrate that MM-STGNN achieves AUROC of 0.79 on both primary and external datasets. Furthermore, MM-STGNN significantly outperforms the current clinical reference standard, LACE+ score (AUROC=0.61), on the primary dataset. For subset populations of patients with heart and vascular disease, our model also outperforms baselines on predicting 30-day readmission (e.g., 3.7 point improvement in AUROC in patients with heart disease). Lastly, qualitative model interpretability analysis indicates that while patients' primary diagnoses were not explicitly used to train the model, node features crucial for model prediction directly reflect patients' primary diagnoses. Importantly, our MM-STGNN is agnostic to node feature modalities and could be utilized to integrate multimodal data for triaging patients in various downstream resource allocation tasks.
Domino: Discovering Systematic Errors with Cross-Modal Embeddings
Apr 11, 2022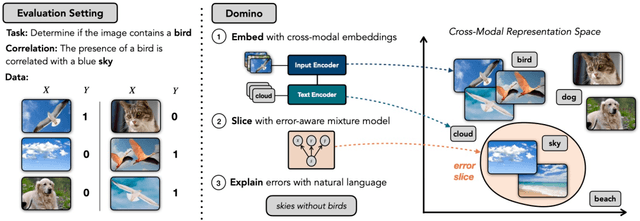
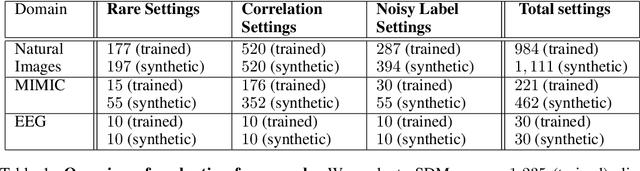
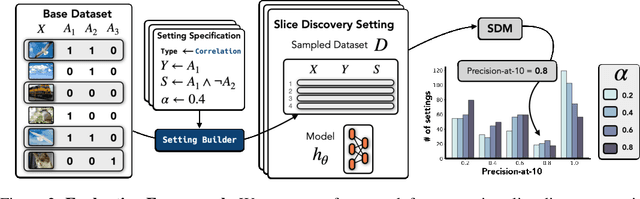
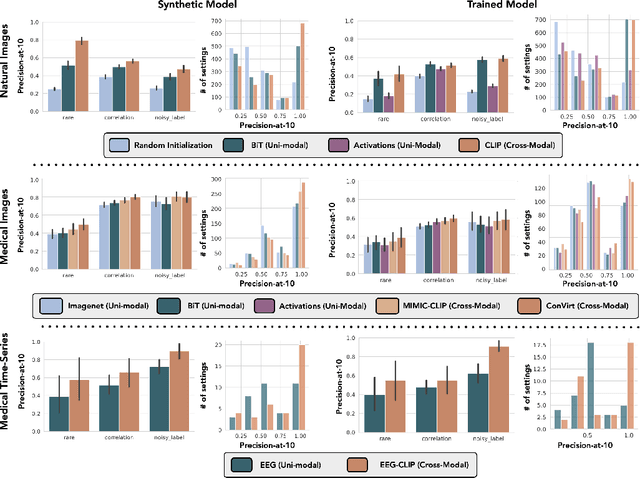
Abstract:Machine learning models that achieve high overall accuracy often make systematic errors on important subsets (or slices) of data. Identifying underperforming slices is particularly challenging when working with high-dimensional inputs (e.g. images, audio), where important slices are often unlabeled. In order to address this issue, recent studies have proposed automated slice discovery methods (SDMs), which leverage learned model representations to mine input data for slices on which a model performs poorly. To be useful to a practitioner, these methods must identify slices that are both underperforming and coherent (i.e. united by a human-understandable concept). However, no quantitative evaluation framework currently exists for rigorously assessing SDMs with respect to these criteria. Additionally, prior qualitative evaluations have shown that SDMs often identify slices that are incoherent. In this work, we address these challenges by first designing a principled evaluation framework that enables a quantitative comparison of SDMs across 1,235 slice discovery settings in three input domains (natural images, medical images, and time-series data). Then, motivated by the recent development of powerful cross-modal representation learning approaches, we present Domino, an SDM that leverages cross-modal embeddings and a novel error-aware mixture model to discover and describe coherent slices. We find that Domino accurately identifies 36% of the 1,235 slices in our framework - a 12 percentage point improvement over prior methods. Further, Domino is the first SDM that can provide natural language descriptions of identified slices, correctly generating the exact name of the slice in 35% of settings.
OncoNet: Weakly Supervised Siamese Network to automate cancer treatment response assessment between longitudinal FDG PET/CT examinations
Aug 03, 2021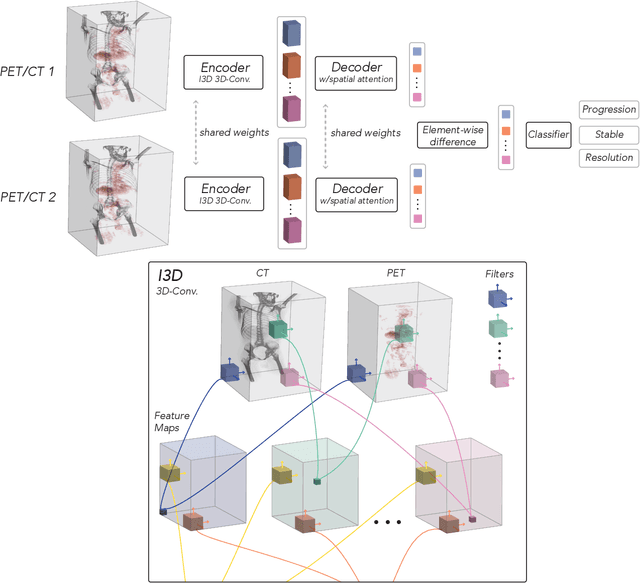

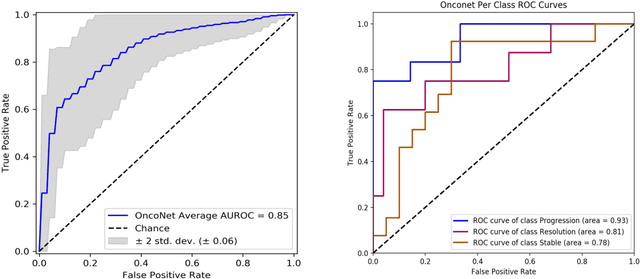
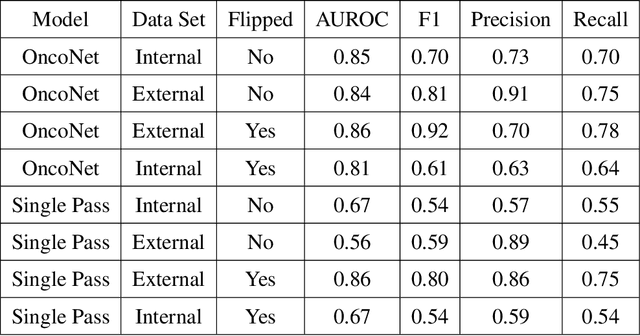
Abstract:FDG PET/CT imaging is a resource intensive examination critical for managing malignant disease and is particularly important for longitudinal assessment during therapy. Approaches to automate longtudinal analysis present many challenges including lack of available longitudinal datasets, managing complex large multimodal imaging examinations, and need for detailed annotations for traditional supervised machine learning. In this work we develop OncoNet, novel machine learning algorithm that assesses treatment response from a 1,954 pairs of sequential FDG PET/CT exams through weak supervision using the standard uptake values (SUVmax) in associated radiology reports. OncoNet demonstrates an AUROC of 0.86 and 0.84 on internal and external institution test sets respectively for determination of change between scans while also showing strong agreement to clinical scoring systems with a kappa score of 0.8. We also curated a dataset of 1,954 paired FDG PET/CT exams designed for response assessment for the broader machine learning in healthcare research community. Automated assessment of radiographic response from FDG PET/CT with OncoNet could provide clinicians with a valuable tool to rapidly and consistently interpret change over time in longitudinal multi-modal imaging exams.
Inaccurate Supervision of Neural Networks with Incorrect Labels: Application to Epilepsy
Dec 01, 2020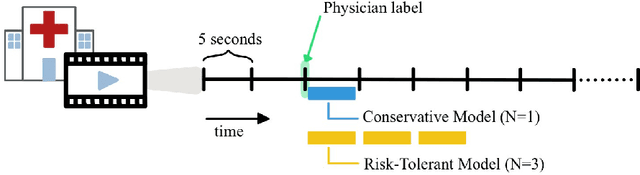
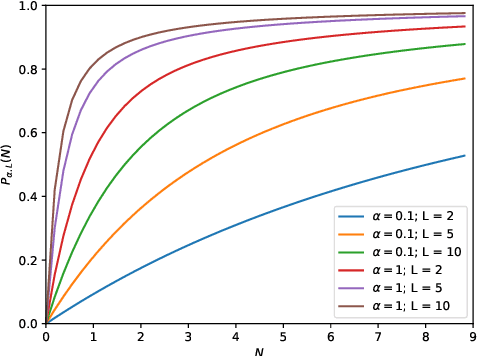
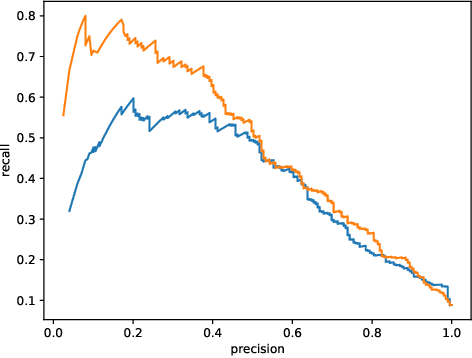
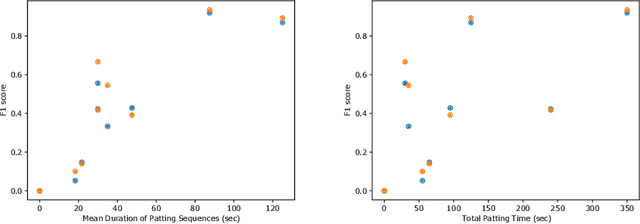
Abstract:This work describes multiple weak supervision strategies for video processing with neural networks in the context of seizure detection. To study seizure onset, we have designed automated methods to detect seizures from electroencephalography (EEG), a modality used for recording electrical brain activity. However, the EEG signal alone is sometimes not enough for existing detection methods to discriminate seizure from artifacts having a similar signal on EEG. For example, such artifacts could be triggered by patting, rocking or suctioning in the case of neonates. In this article, we addressed this problem by automatically detecting an example artifact -- patting of neonates -- from continuous video recordings of neonates acquired during clinical routine. We computed frame-to-frame cross-correlation matrices to isolate patterns showing repetitive movements indicative of patting of the patient. Next, a convolutional neural network was trained to classify whether these matrices contained patting events using weak training labels -- noisy labels generated during daily clinical procedure. The labels were considered weak as they were sometimes incorrect. We investigated whether networks trained with more samples, containing more uncertain and weak labels, could achieve a higher performance. Our results showed that, in the case of patting detection, such networks could achieve a higher recall, without sacrificing precision. These networks focused on areas of the cross-correlation matrices that were more meaningful to the task. More generally, our work gives insights into building more accurate models from weakly labelled time sequences.
Ivy: Instrumental Variable Synthesis for Causal Inference
Apr 11, 2020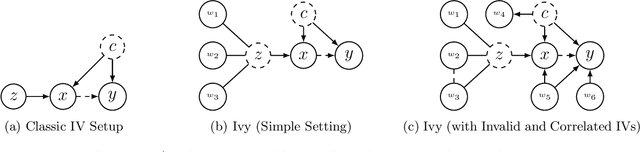
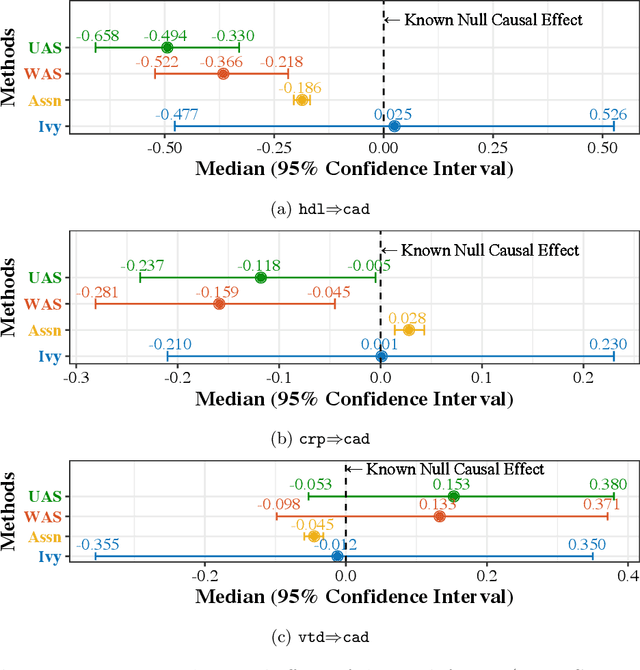
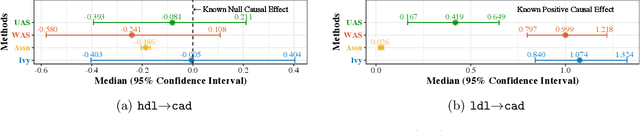
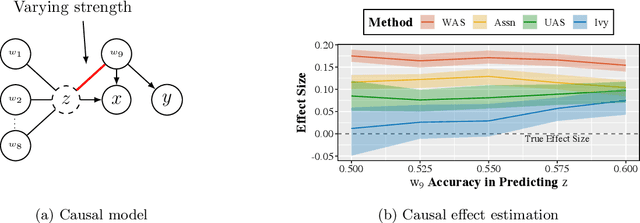
Abstract:A popular way to estimate the causal effect of a variable x on y from observational data is to use an instrumental variable (IV): a third variable z that affects y only through x. The more strongly z is associated with x, the more reliable the estimate is, but such strong IVs are difficult to find. Instead, practitioners combine more commonly available IV candidates---which are not necessarily strong, or even valid, IVs---into a single "summary" that is plugged into causal effect estimators in place of an IV. In genetic epidemiology, such approaches are known as allele scores. Allele scores require strong assumptions---independence and validity of all IV candidates---for the resulting estimate to be reliable. To relax these assumptions, we propose Ivy, a new method to combine IV candidates that can handle correlated and invalid IV candidates in a robust manner. Theoretically, we characterize this robustness, its limits, and its impact on the resulting causal estimates. Empirically, Ivy can correctly identify the directionality of known relationships and is robust against false discovery (median effect size <= 0.025) on three real-world datasets with no causal effects, while allele scores return more biased estimates (median effect size >= 0.118).
Hidden Stratification Causes Clinically Meaningful Failures in Machine Learning for Medical Imaging
Sep 27, 2019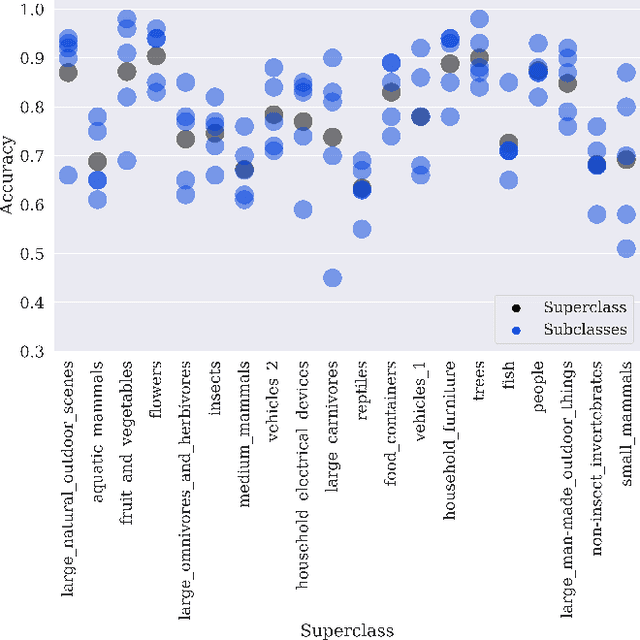
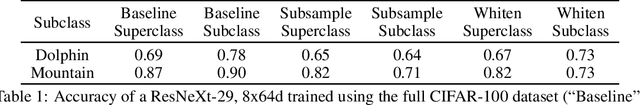
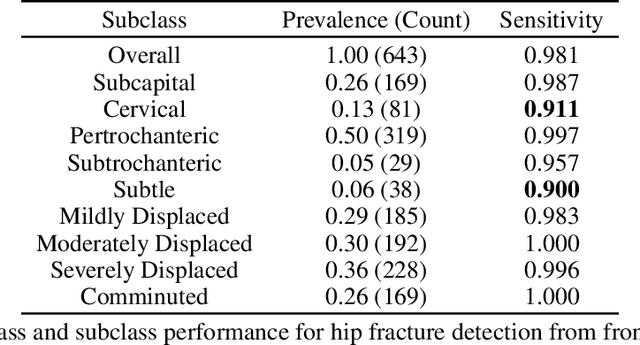
Abstract:Machine learning models for medical image analysis often suffer from poor performance on important subsets of a population that are not identified during training or testing. For example, overall performance of a cancer detection model may be high, but the model still consistently misses a rare but aggressive cancer subtype. We refer to this problem as hidden stratification, and observe that it results from incompletely describing the meaningful variation in a dataset. While hidden stratification can substantially reduce the clinical efficacy of machine learning models, its effects remain difficult to measure. In this work, we assess the utility of several possible techniques for measuring and describing hidden stratification effects, and characterize these effects both on multiple medical imaging datasets and via synthetic experiments on the well-characterised CIFAR-100 benchmark dataset. We find evidence that hidden stratification can occur in unidentified imaging subsets with low prevalence, low label quality, subtle distinguishing features, or spurious correlates, and that it can result in relative performance differences of over 20% on clinically important subsets. Finally, we explore the clinical implications of our findings, and suggest that evaluation of hidden stratification should be a critical component of any machine learning deployment in medical imaging.
Cross-Modal Data Programming Enables Rapid Medical Machine Learning
Mar 26, 2019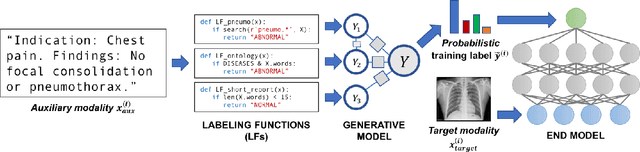
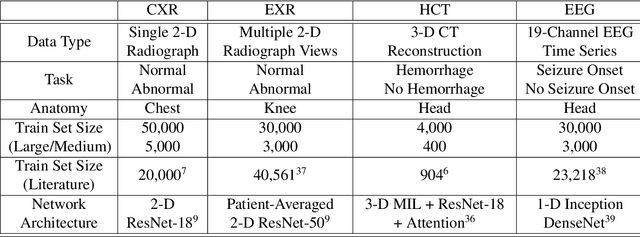
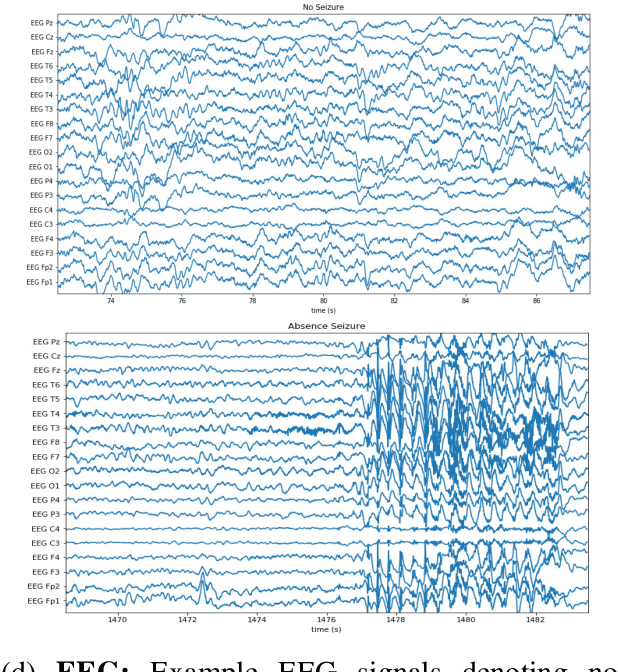
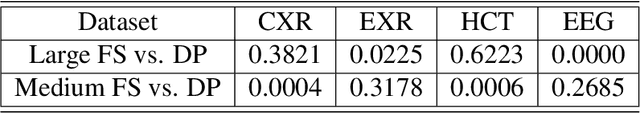
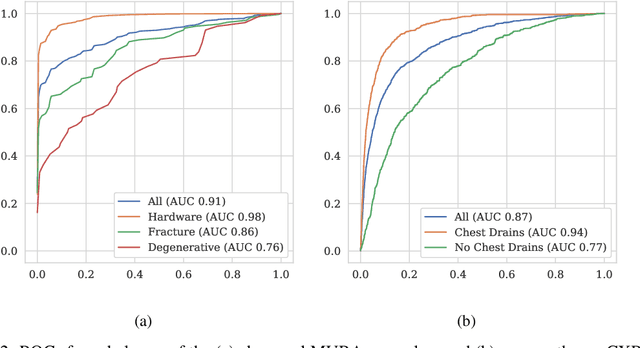
Abstract:Labeling training datasets has become a key barrier to building medical machine learning models. One strategy is to generate training labels programmatically, for example by applying natural language processing pipelines to text reports associated with imaging studies. We propose cross-modal data programming, which generalizes this intuitive strategy in a theoretically-grounded way that enables simpler, clinician-driven input, reduces required labeling time, and improves with additional unlabeled data. In this approach, clinicians generate training labels for models defined over a target modality (e.g. images or time series) by writing rules over an auxiliary modality (e.g. text reports). The resulting technical challenge consists of estimating the accuracies and correlations of these rules; we extend a recent unsupervised generative modeling technique to handle this cross-modal setting in a provably consistent way. Across four applications in radiography, computed tomography, and electroencephalography, and using only several hours of clinician time, our approach matches or exceeds the efficacy of physician-months of hand-labeling with statistical significance, demonstrating a fundamentally faster and more flexible way of building machine learning models in medicine.
Predicting US State-Level Agricultural Sentiment as a Measure of Food Security with Tweets from Farming Communities
Feb 13, 2019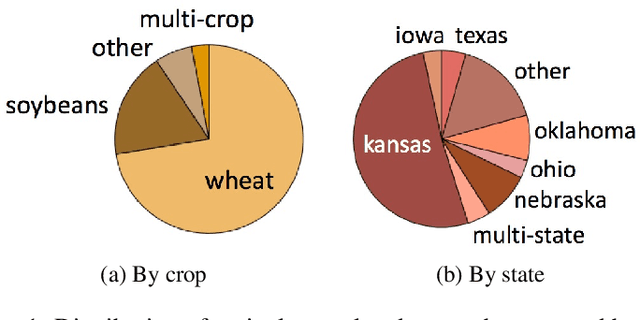
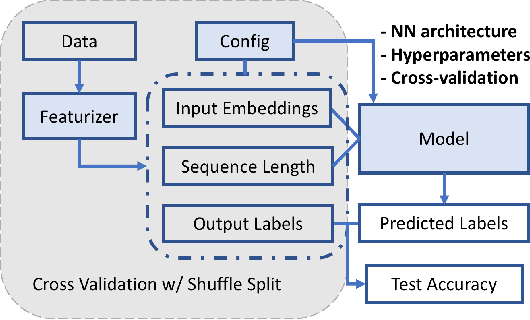
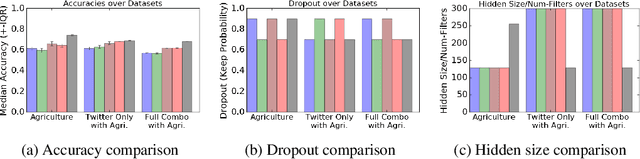
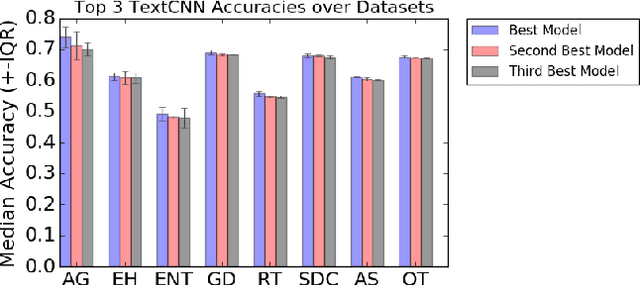
Abstract:The ability to obtain accurate food security metrics in developing areas where relevant data can be sparse is critically important for policy makers tasked with implementing food aid programs. As a result, a great deal of work has been dedicated to predicting important food security metrics such as annual crop yields using a variety of methods including simulation, remote sensing, weather models, and human expert input. As a complement to existing techniques in crop yield prediction, this work develops neural network models for predicting the sentiment of Twitter feeds from farming communities. Specifically, we investigate the potential of both direct learning on a small dataset of agriculturally-relevant tweets and transfer learning from larger, well-labeled sentiment datasets from other domains (e.g.~politics) to accurately predict agricultural sentiment, which we hope would ultimately serve as a useful crop yield predictor. We find that direct learning from small, relevant datasets outperforms transfer learning from large, fully-labeled datasets, that convolutional neural networks broadly outperform recurrent neural networks on Twitter sentiment classification, and that these models perform substantially less well on ternary sentiment problems characteristic of practical settings than on binary problems often found in the literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge