Hersh Sagreiya
Evidence Is All You Need: Ordering Imaging Studies via Language Model Alignment with the ACR Appropriateness Criteria
Sep 27, 2024



Abstract:Diagnostic imaging studies are an increasingly important component of the workup and management of acutely presenting patients. However, ordering appropriate imaging studies according to evidence-based medical guidelines is a challenging task with a high degree of variability between healthcare providers. To address this issue, recent work has investigated if generative AI and large language models can be leveraged to help clinicians order relevant imaging studies for patients. However, it is challenging to ensure that these tools are correctly aligned with medical guidelines, such as the American College of Radiology's Appropriateness Criteria (ACR AC). In this study, we introduce a framework to intelligently leverage language models by recommending imaging studies for patient cases that are aligned with evidence-based guidelines. We make available a novel dataset of patient "one-liner" scenarios to power our experiments, and optimize state-of-the-art language models to achieve an accuracy on par with clinicians in image ordering. Finally, we demonstrate that our language model-based pipeline can be used as intelligent assistants by clinicians to support image ordering workflows and improve the accuracy of imaging study ordering according to the ACR AC. Our work demonstrates and validates a strategy to leverage AI-based software to improve trustworthy clinical decision making in alignment with expert evidence-based guidelines.
Machine learning-based algorithms for at-home respiratory disease monitoring and respiratory assessment
Sep 05, 2024



Abstract:Respiratory diseases impose a significant burden on global health, with current diagnostic and management practices primarily reliant on specialist clinical testing. This work aims to develop machine learning-based algorithms to facilitate at-home respiratory disease monitoring and assessment for patients undergoing continuous positive airway pressure (CPAP) therapy. Data were collected from 30 healthy adults, encompassing respiratory pressure, flow, and dynamic thoraco-abdominal circumferential measurements under three breathing conditions: normal, panting, and deep breathing. Various machine learning models, including the random forest classifier, logistic regression, and support vector machine (SVM), were trained to predict breathing types. The random forest classifier demonstrated the highest accuracy, particularly when incorporating breathing rate as a feature. These findings support the potential of AI-driven respiratory monitoring systems to transition respiratory assessments from clinical settings to home environments, enhancing accessibility and patient autonomy. Future work involves validating these models with larger, more diverse populations and exploring additional machine learning techniques.
CopilotCAD: Empowering Radiologists with Report Completion Models and Quantitative Evidence from Medical Image Foundation Models
Apr 11, 2024



Abstract:Computer-aided diagnosis systems hold great promise to aid radiologists and clinicians in radiological clinical practice and enhance diagnostic accuracy and efficiency. However, the conventional systems primarily focus on delivering diagnostic results through text report generation or medical image classification, positioning them as standalone decision-makers rather than helpers and ignoring radiologists' expertise. This study introduces an innovative paradigm to create an assistive co-pilot system for empowering radiologists by leveraging Large Language Models (LLMs) and medical image analysis tools. Specifically, we develop a collaborative framework to integrate LLMs and quantitative medical image analysis results generated by foundation models with radiologists in the loop, achieving efficient and safe generation of radiology reports and effective utilization of computational power of AI and the expertise of medical professionals. This approach empowers radiologists to generate more precise and detailed diagnostic reports, enhancing patient outcomes while reducing the burnout of clinicians. Our methodology underscores the potential of AI as a supportive tool in medical diagnostics, promoting a harmonious integration of technology and human expertise to advance the field of radiology.
Learning-Based Radiomic Prediction of Type 2 Diabetes Mellitus Using Image-Derived Phenotypes
Sep 20, 2022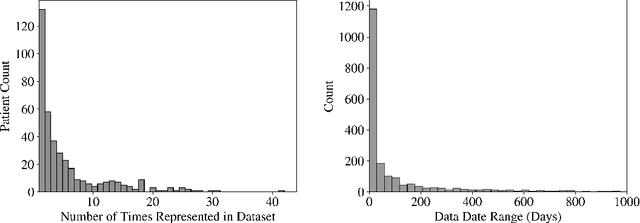
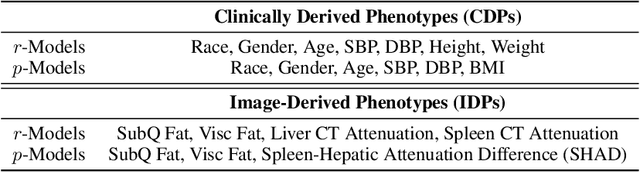
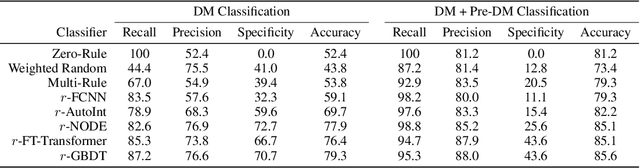
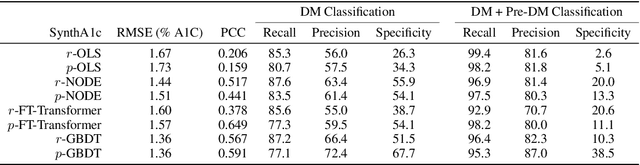
Abstract:Early diagnosis of Type 2 Diabetes Mellitus (T2DM) is crucial to enable timely therapeutic interventions and lifestyle modifications. As medical imaging data become more widely available for many patient populations, we sought to investigate whether image-derived phenotypic data could be leveraged in tabular learning classifier models to predict T2DM incidence without the use of invasive blood lab measurements. We show that both neural network and decision tree models that use image-derived phenotypes can predict patient T2DM status with recall scores as high as 87.6%. We also propose the novel use of these same architectures as 'SynthA1c encoders' that are able to output interpretable values mimicking blood hemoglobin A1C empirical lab measurements. Finally, we demonstrate that T2DM risk prediction model sensitivity to small perturbations in input vector components can be used to predict performance on covariates sampled from previously unseen patient populations.
Cross-Modal Data Programming Enables Rapid Medical Machine Learning
Mar 26, 2019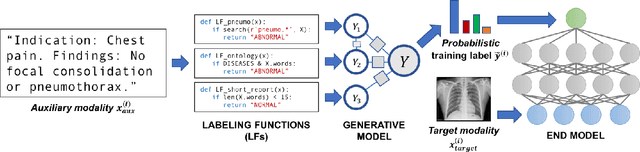
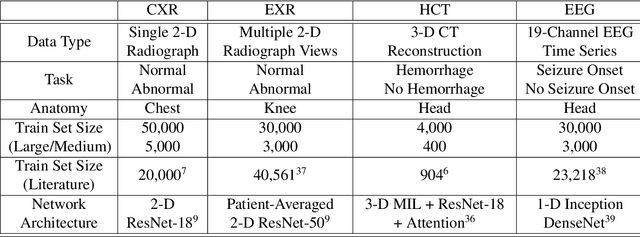
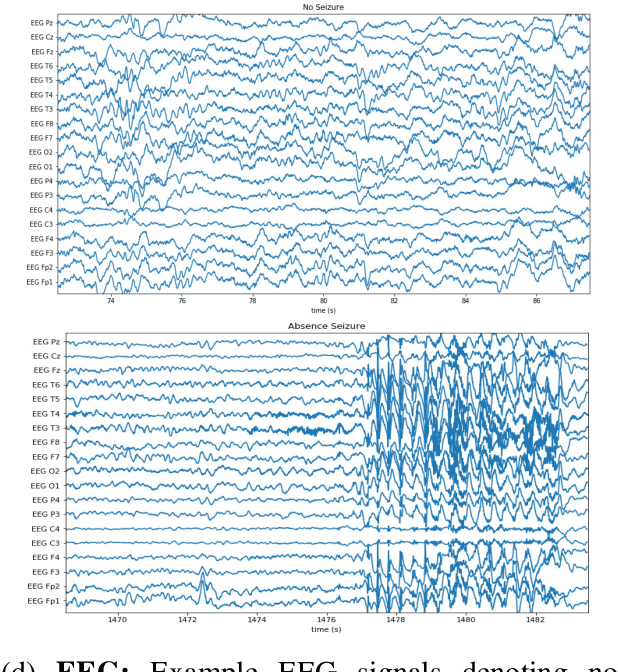
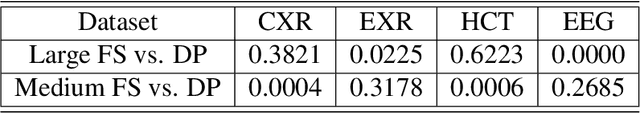
Abstract:Labeling training datasets has become a key barrier to building medical machine learning models. One strategy is to generate training labels programmatically, for example by applying natural language processing pipelines to text reports associated with imaging studies. We propose cross-modal data programming, which generalizes this intuitive strategy in a theoretically-grounded way that enables simpler, clinician-driven input, reduces required labeling time, and improves with additional unlabeled data. In this approach, clinicians generate training labels for models defined over a target modality (e.g. images or time series) by writing rules over an auxiliary modality (e.g. text reports). The resulting technical challenge consists of estimating the accuracies and correlations of these rules; we extend a recent unsupervised generative modeling technique to handle this cross-modal setting in a provably consistent way. Across four applications in radiography, computed tomography, and electroencephalography, and using only several hours of clinician time, our approach matches or exceeds the efficacy of physician-months of hand-labeling with statistical significance, demonstrating a fundamentally faster and more flexible way of building machine learning models in medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge