Allison Chae
Evidence Is All You Need: Ordering Imaging Studies via Language Model Alignment with the ACR Appropriateness Criteria
Sep 27, 2024



Abstract:Diagnostic imaging studies are an increasingly important component of the workup and management of acutely presenting patients. However, ordering appropriate imaging studies according to evidence-based medical guidelines is a challenging task with a high degree of variability between healthcare providers. To address this issue, recent work has investigated if generative AI and large language models can be leveraged to help clinicians order relevant imaging studies for patients. However, it is challenging to ensure that these tools are correctly aligned with medical guidelines, such as the American College of Radiology's Appropriateness Criteria (ACR AC). In this study, we introduce a framework to intelligently leverage language models by recommending imaging studies for patient cases that are aligned with evidence-based guidelines. We make available a novel dataset of patient "one-liner" scenarios to power our experiments, and optimize state-of-the-art language models to achieve an accuracy on par with clinicians in image ordering. Finally, we demonstrate that our language model-based pipeline can be used as intelligent assistants by clinicians to support image ordering workflows and improve the accuracy of imaging study ordering according to the ACR AC. Our work demonstrates and validates a strategy to leverage AI-based software to improve trustworthy clinical decision making in alignment with expert evidence-based guidelines.
Learning-Based Radiomic Prediction of Type 2 Diabetes Mellitus Using Image-Derived Phenotypes
Sep 20, 2022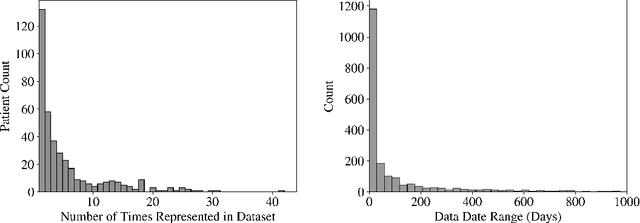
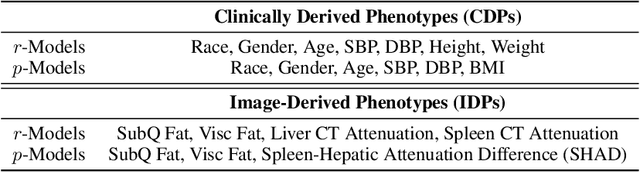
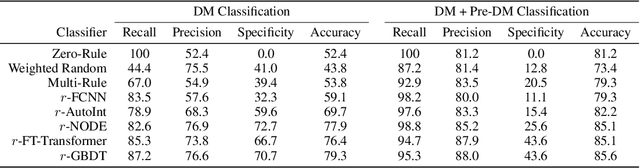
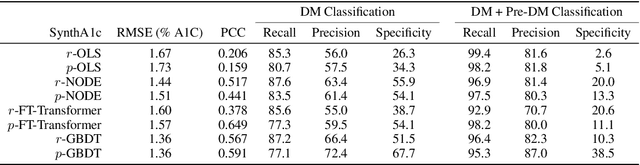
Abstract:Early diagnosis of Type 2 Diabetes Mellitus (T2DM) is crucial to enable timely therapeutic interventions and lifestyle modifications. As medical imaging data become more widely available for many patient populations, we sought to investigate whether image-derived phenotypic data could be leveraged in tabular learning classifier models to predict T2DM incidence without the use of invasive blood lab measurements. We show that both neural network and decision tree models that use image-derived phenotypes can predict patient T2DM status with recall scores as high as 87.6%. We also propose the novel use of these same architectures as 'SynthA1c encoders' that are able to output interpretable values mimicking blood hemoglobin A1C empirical lab measurements. Finally, we demonstrate that T2DM risk prediction model sensitivity to small perturbations in input vector components can be used to predict performance on covariates sampled from previously unseen patient populations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge