James C. Gee
The LUMirage: An independent evaluation of zero-shot performance in the LUMIR challenge
Dec 17, 2025Abstract:The LUMIR challenge represents an important benchmark for evaluating deformable image registration methods on large-scale neuroimaging data. While the challenge demonstrates that modern deep learning methods achieve competitive accuracy on T1-weighted MRI, it also claims exceptional zero-shot generalization to unseen contrasts and resolutions, assertions that contradict established understanding of domain shift in deep learning. In this paper, we perform an independent re-evaluation of these zero-shot claims using rigorous evaluation protocols while addressing potential sources of instrumentation bias. Our findings reveal a more nuanced picture: (1) deep learning methods perform comparably to iterative optimization on in-distribution T1w images and even on human-adjacent species (macaque), demonstrating improved task understanding; (2) however, performance degrades significantly on out-of-distribution contrasts (T2, T2*, FLAIR), with Cohen's d scores ranging from 0.7-1.5, indicating substantial practical impact on downstream clinical workflows; (3) deep learning methods face scalability limitations on high-resolution data, failing to run on 0.6 mm isotropic images, while iterative methods benefit from increased resolution; and (4) deep methods exhibit high sensitivity to preprocessing choices. These results align with the well-established literature on domain shift and suggest that claims of universal zero-shot superiority require careful scrutiny. We advocate for evaluation protocols that reflect practical clinical and research workflows rather than conditions that may inadvertently favor particular method classes.
Diversity By Design: Leveraging Distribution Matching for Offline Model-Based Optimization
Jan 30, 2025

Abstract:The goal of offline model-based optimization (MBO) is to propose new designs that maximize a reward function given only an offline dataset. However, an important desiderata is to also propose a diverse set of final candidates that capture many optimal and near-optimal design configurations. We propose Diversity in Adversarial Model-based Optimization (DynAMO) as a novel method to introduce design diversity as an explicit objective into any MBO problem. Our key insight is to formulate diversity as a distribution matching problem where the distribution of generated designs captures the inherent diversity contained within the offline dataset. Extensive experiments spanning multiple scientific domains show that DynAMO can be used with common optimization methods to significantly improve the diversity of proposed designs while still discovering high-quality candidates.
Evidence Is All You Need: Ordering Imaging Studies via Language Model Alignment with the ACR Appropriateness Criteria
Sep 27, 2024



Abstract:Diagnostic imaging studies are an increasingly important component of the workup and management of acutely presenting patients. However, ordering appropriate imaging studies according to evidence-based medical guidelines is a challenging task with a high degree of variability between healthcare providers. To address this issue, recent work has investigated if generative AI and large language models can be leveraged to help clinicians order relevant imaging studies for patients. However, it is challenging to ensure that these tools are correctly aligned with medical guidelines, such as the American College of Radiology's Appropriateness Criteria (ACR AC). In this study, we introduce a framework to intelligently leverage language models by recommending imaging studies for patient cases that are aligned with evidence-based guidelines. We make available a novel dataset of patient "one-liner" scenarios to power our experiments, and optimize state-of-the-art language models to achieve an accuracy on par with clinicians in image ordering. Finally, we demonstrate that our language model-based pipeline can be used as intelligent assistants by clinicians to support image ordering workflows and improve the accuracy of imaging study ordering according to the ACR AC. Our work demonstrates and validates a strategy to leverage AI-based software to improve trustworthy clinical decision making in alignment with expert evidence-based guidelines.
Deep Implicit Optimization for Robust and Flexible Image Registration
Jun 11, 2024



Abstract:Deep Learning in Image Registration (DLIR) methods have been tremendously successful in image registration due to their speed and ability to incorporate weak label supervision at training time. However, DLIR methods forego many of the benefits of classical optimization-based methods. The functional nature of deep networks do not guarantee that the predicted transformation is a local minima of the registration objective, the representation of the transformation (displacement/velocity field/affine) is fixed, and the networks are not robust to domain shift. Our method aims to bridge this gap between classical and learning methods by incorporating optimization as a layer in a deep network. A deep network is trained to predict multi-scale dense feature images that are registered using a black box iterative optimization solver. This optimal warp is then used to minimize image and label alignment errors. By implicitly differentiating end-to-end through an iterative optimization solver, our learned features are registration and label-aware, and the warp functions are guaranteed to be local minima of the registration objective in the feature space. Our framework shows excellent performance on in-domain datasets, and is agnostic to domain shift such as anisotropy and varying intensity profiles. For the first time, our method allows switching between arbitrary transformation representations (free-form to diffeomorphic) at test time with zero retraining. End-to-end feature learning also facilitates interpretability of features, and out-of-the-box promptability using additional label-fidelity terms at inference.
A Textbook Remedy for Domain Shifts: Knowledge Priors for Medical Image Analysis
May 23, 2024



Abstract:While deep networks have achieved broad success in analyzing natural images, when applied to medical scans, they often fail in unexcepted situations. We investigate this challenge and focus on model sensitivity to domain shifts, such as data sampled from different hospitals or data confounded by demographic variables such as sex, race, etc, in the context of chest X-rays and skin lesion images. A key finding we show empirically is that existing visual backbones lack an appropriate prior from the architecture for reliable generalization in these settings. Taking inspiration from medical training, we propose giving deep networks a prior grounded in explicit medical knowledge communicated in natural language. To this end, we introduce Knowledge-enhanced Bottlenecks (KnoBo), a class of concept bottleneck models that incorporates knowledge priors that constrain it to reason with clinically relevant factors found in medical textbooks or PubMed. KnoBo uses retrieval-augmented language models to design an appropriate concept space paired with an automatic training procedure for recognizing the concept. We evaluate different resources of knowledge and recognition architectures on a broad range of domain shifts across 20 datasets. In our comprehensive evaluation with two imaging modalities, KnoBo outperforms fine-tuned models on confounded datasets by 32.4% on average. Finally, evaluations reveal that PubMed is a promising resource for making medical models less sensitive to domain shift, outperforming other resources on both diversity of information and final prediction performance.
Neural Ordinary Differential Equation based Sequential Image Registration for Dynamic Characterization
Apr 02, 2024



Abstract:Deformable image registration (DIR) is crucial in medical image analysis, enabling the exploration of biological dynamics such as organ motions and longitudinal changes in imaging. Leveraging Neural Ordinary Differential Equations (ODE) for registration, this extension work discusses how this framework can aid in the characterization of sequential biological processes. Utilizing the Neural ODE's ability to model state derivatives with neural networks, our Neural Ordinary Differential Equation Optimization-based (NODEO) framework considers voxels as particles within a dynamic system, defining deformation fields through the integration of neural differential equations. This method learns dynamics directly from data, bypassing the need for physical priors, making it exceptionally suitable for medical scenarios where such priors are unavailable or inapplicable. Consequently, the framework can discern underlying dynamics and use sequence data to regularize the transformation trajectory. We evaluated our framework on two clinical datasets: one for cardiac motion tracking and another for longitudinal brain MRI analysis. Demonstrating its efficacy in both 2D and 3D imaging scenarios, our framework offers flexibility and model agnosticism, capable of managing image sequences and facilitating label propagation throughout these sequences. This study provides a comprehensive understanding of how the Neural ODE-based framework uniquely benefits the image registration challenge.
FireANTs: Adaptive Riemannian Optimization for Multi-Scale Diffeomorphic Registration
Apr 01, 2024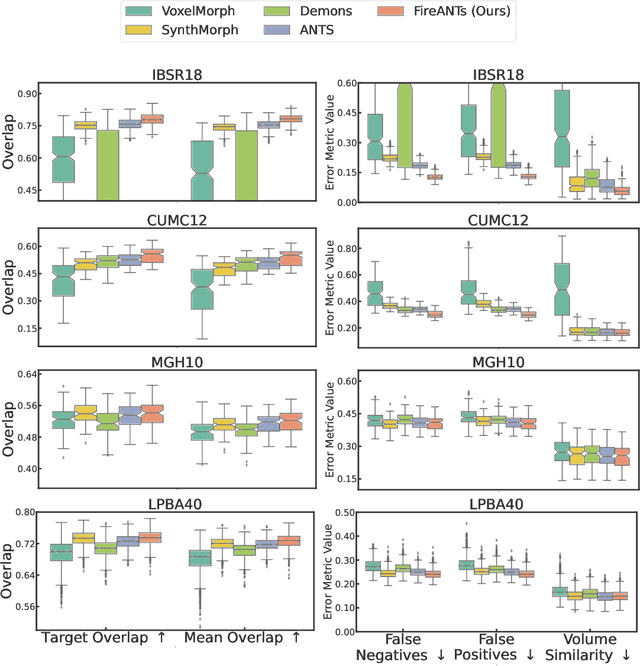
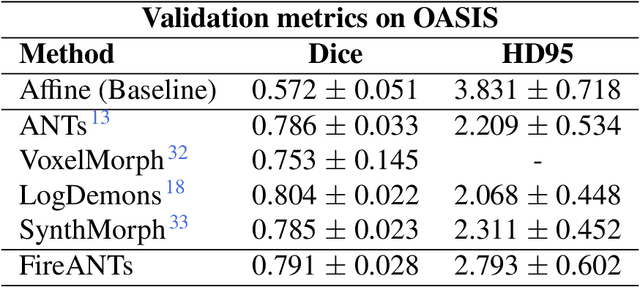
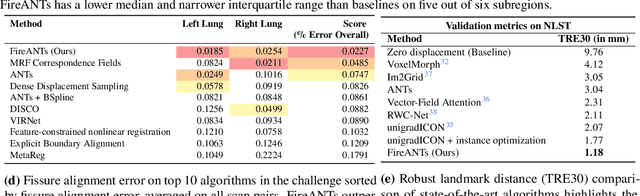
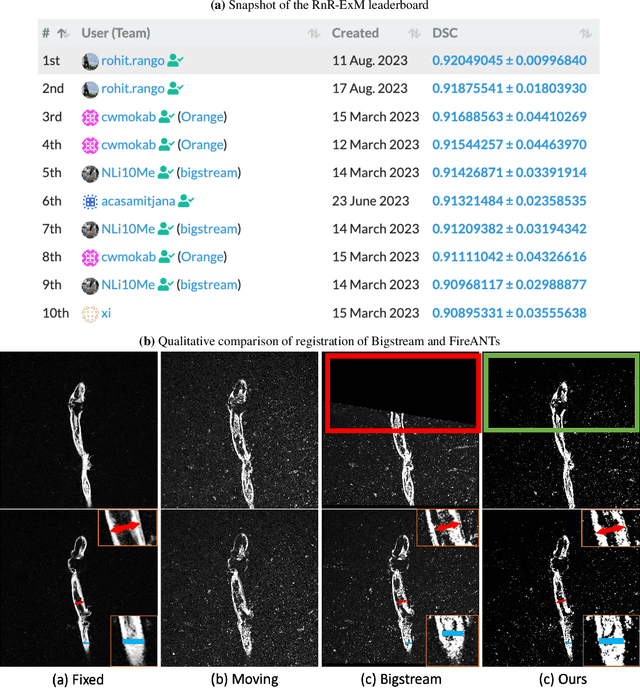
Abstract:Diffeomorphic Image Registration is a critical part of the analysis in various imaging modalities and downstream tasks like image translation, segmentation, and atlas building. Registration algorithms based on optimization have stood the test of time in terms of accuracy, reliability, and robustness across a wide spectrum of modalities and acquisition settings. However, these algorithms converge slowly, are prohibitively expensive to run, and their usage requires a steep learning curve, limiting their scalability to larger clinical and scientific studies. In this paper, we develop multi-scale Adaptive Riemannian Optimization algorithms for diffeomorphic image registration. We demonstrate compelling improvements on image registration across a spectrum of modalities and anatomies by measuring structural and landmark overlap of the registered image volumes. Our proposed framework leads to a consistent improvement in performance, and from 300x up to 2000x speedup over existing algorithms. Our modular library design makes it easy to use and allows customization via user-defined cost functions.
A Concept-based Interpretable Model for the Diagnosis of Choroid Neoplasias using Multimodal Data
Mar 08, 2024Abstract:Diagnosing rare diseases presents a common challenge in clinical practice, necessitating the expertise of specialists for accurate identification. The advent of machine learning offers a promising solution, while the development of such technologies is hindered by the scarcity of data on rare conditions and the demand for models that are both interpretable and trustworthy in a clinical context. Interpretable AI, with its capacity for human-readable outputs, can facilitate validation by clinicians and contribute to medical education. In the current work, we focus on choroid neoplasias, the most prevalent form of eye cancer in adults, albeit rare with 5.1 per million. We built the so-far largest dataset consisting of 750 patients, incorporating three distinct imaging modalities collected from 2004 to 2022. Our work introduces a concept-based interpretable model that distinguishes between three types of choroidal tumors, integrating insights from domain experts via radiological reports. Remarkably, this model not only achieves an F1 score of 0.91, rivaling that of black-box models, but also boosts the diagnostic accuracy of junior doctors by 42%. This study highlights the significant potential of interpretable machine learning in improving the diagnosis of rare diseases, laying a groundwork for future breakthroughs in medical AI that could tackle a wider array of complex health scenarios.
Generative Adversarial Bayesian Optimization for Surrogate Objectives
Feb 09, 2024Abstract:Offline model-based policy optimization seeks to optimize a learned surrogate objective function without querying the true oracle objective during optimization. However, inaccurate surrogate model predictions are frequently encountered along the optimization trajectory. To address this limitation, we propose generative adversarial Bayesian optimization (GABO) using adaptive source critic regularization, a task-agnostic framework for Bayesian optimization that employs a Lipschitz-bounded source critic model to constrain the optimization trajectory to regions where the surrogate function is reliable. We show that under certain assumptions for the continuous input space prior, our algorithm dynamically adjusts the strength of the source critic regularization. GABO outperforms existing baselines on a number of different offline optimization tasks across a variety of scientific domains. Our code is available at https://github.com/michael-s-yao/gabo
Towards Establishing Dense Correspondence on Multiview Coronary Angiography: From Point-to-Point to Curve-to-Curve Query Matching
Dec 18, 2023Abstract:Coronary angiography is the gold standard imaging technique for studying and diagnosing coronary artery disease. However, the resulting 2D X-ray projections lose 3D information and exhibit visual ambiguities. In this work, we aim to establish dense correspondence in multi-view angiography, serving as a fundamental basis for various clinical applications and downstream tasks. To overcome the challenge of unavailable annotated data, we designed a data simulation pipeline using 3D Coronary Computed Tomography Angiography (CCTA). We formulated the problem of dense correspondence estimation as a query matching task over all points of interest in the given views. We established point-to-point query matching and advanced it to curve-to-curve correspondence, significantly reducing errors by minimizing ambiguity and improving topological awareness. The method was evaluated on a set of 1260 image pairs from different views across 8 clinically relevant angulation groups, demonstrating compelling results and indicating the feasibility of establishing dense correspondence in multi-view angiography.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge