Matthew Lungren
CancerGUIDE: Cancer Guideline Understanding via Internal Disagreement Estimation
Sep 09, 2025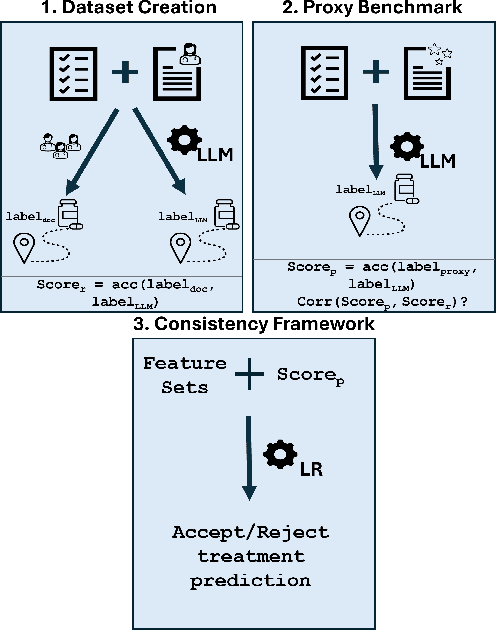
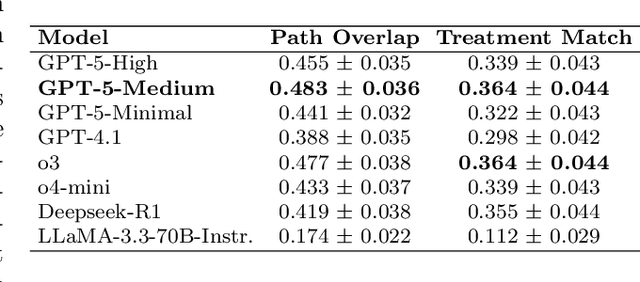
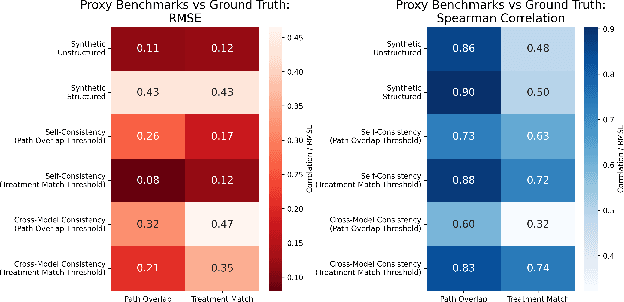
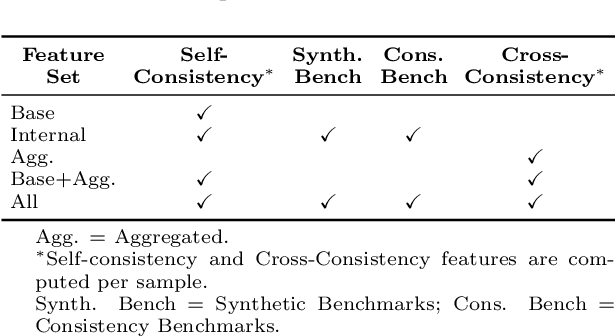
Abstract:The National Comprehensive Cancer Network (NCCN) provides evidence-based guidelines for cancer treatment. Translating complex patient presentations into guideline-compliant treatment recommendations is time-intensive, requires specialized expertise, and is prone to error. Advances in large language model (LLM) capabilities promise to reduce the time required to generate treatment recommendations and improve accuracy. We present an LLM agent-based approach to automatically generate guideline-concordant treatment trajectories for patients with non-small cell lung cancer (NSCLC). Our contributions are threefold. First, we construct a novel longitudinal dataset of 121 cases of NSCLC patients that includes clinical encounters, diagnostic results, and medical histories, each expertly annotated with the corresponding NCCN guideline trajectories by board-certified oncologists. Second, we demonstrate that existing LLMs possess domain-specific knowledge that enables high-quality proxy benchmark generation for both model development and evaluation, achieving strong correlation (Spearman coefficient r=0.88, RMSE = 0.08) with expert-annotated benchmarks. Third, we develop a hybrid approach combining expensive human annotations with model consistency information to create both the agent framework that predicts the relevant guidelines for a patient, as well as a meta-classifier that verifies prediction accuracy with calibrated confidence scores for treatment recommendations (AUROC=0.800), a critical capability for communicating the accuracy of outputs, custom-tailoring tradeoffs in performance, and supporting regulatory compliance. This work establishes a framework for clinically viable LLM-based guideline adherence systems that balance accuracy, interpretability, and regulatory requirements while reducing annotation costs, providing a scalable pathway toward automated clinical decision support.
Multi-Modal Mamba Modeling for Survival Prediction (M4Survive): Adapting Joint Foundation Model Representations
Mar 13, 2025Abstract:Accurate survival prediction in oncology requires integrating diverse imaging modalities to capture the complex interplay of tumor biology. Traditional single-modality approaches often fail to leverage the complementary insights provided by radiological and pathological assessments. In this work, we introduce M4Survive (Multi-Modal Mamba Modeling for Survival Prediction), a novel framework that learns joint foundation model representations using efficient adapter networks. Our approach dynamically fuses heterogeneous embeddings from a foundation model repository (e.g., MedImageInsight, BiomedCLIP, Prov-GigaPath, UNI2-h), creating a correlated latent space optimized for survival risk estimation. By leveraging Mamba-based adapters, M4Survive enables efficient multi-modal learning while preserving computational efficiency. Experimental evaluations on benchmark datasets demonstrate that our approach outperforms both unimodal and traditional static multi-modal baselines in survival prediction accuracy. This work underscores the potential of foundation model-driven multi-modal fusion in advancing precision oncology and predictive analytics.
MedImageInsight: An Open-Source Embedding Model for General Domain Medical Imaging
Oct 09, 2024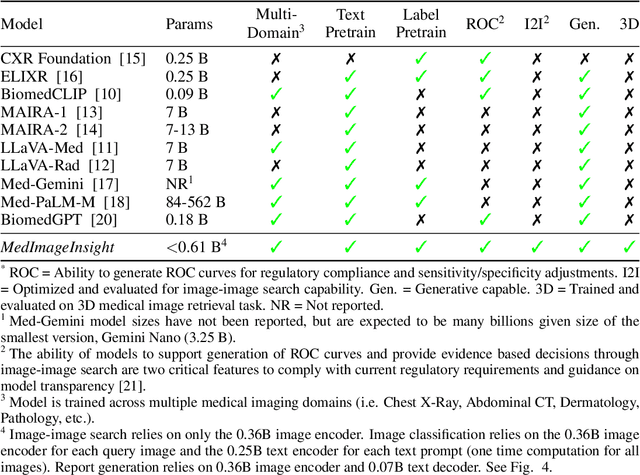
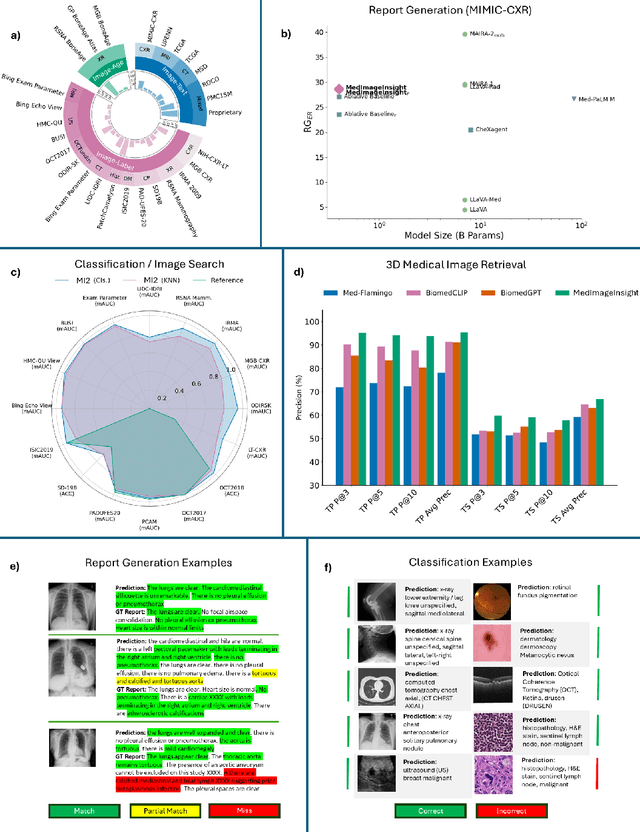
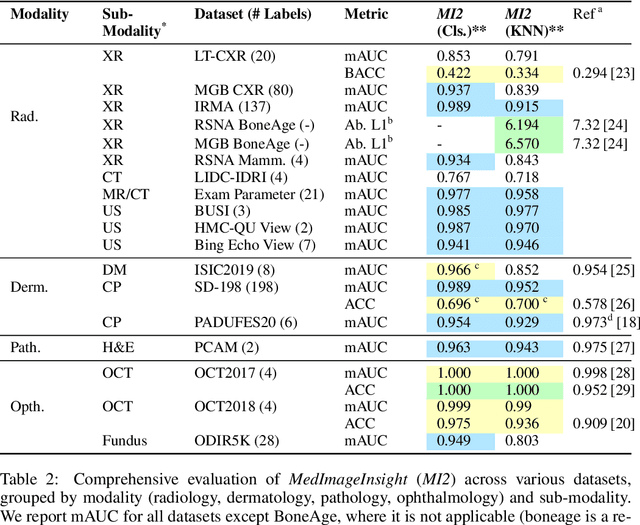
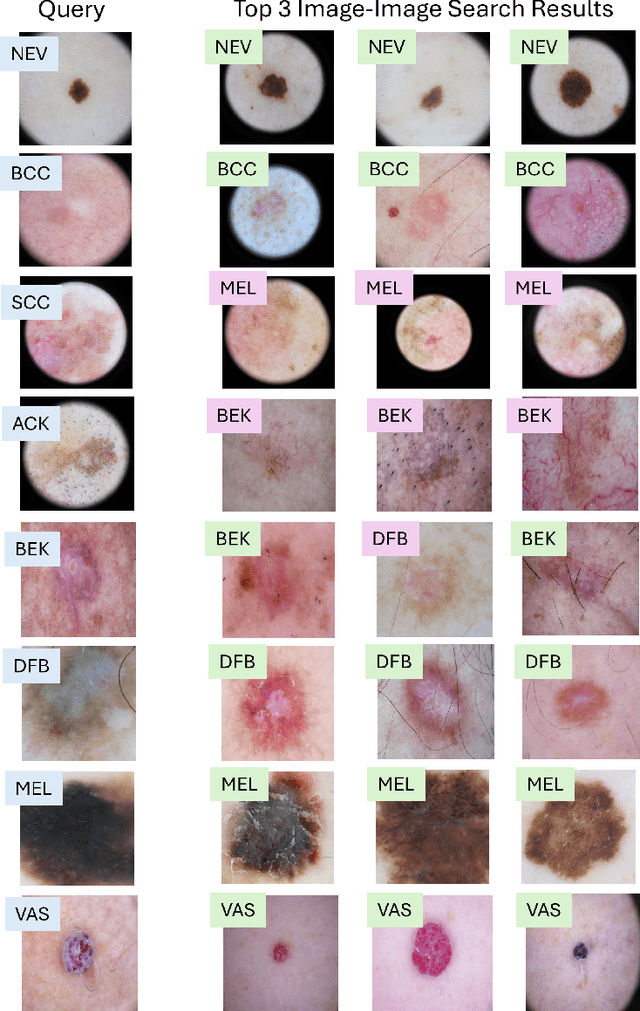
Abstract:In this work, we present MedImageInsight, an open-source medical imaging embedding model. MedImageInsight is trained on medical images with associated text and labels across a diverse collection of domains, including X-Ray, CT, MRI, dermoscopy, OCT, fundus photography, ultrasound, histopathology, and mammography. Rigorous evaluations demonstrate MedImageInsight's ability to achieve state-of-the-art (SOTA) or human expert level performance across classification, image-image search, and fine-tuning tasks. Specifically, on public datasets, MedImageInsight achieves SOTA in CT 3D medical image retrieval, as well as SOTA in disease classification and search for chest X-ray, dermatology, and OCT imaging. Furthermore, MedImageInsight achieves human expert performance in bone age estimation (on both public and partner data), as well as AUC above 0.9 in most other domains. When paired with a text decoder, MedImageInsight achieves near SOTA level single image report findings generation with less than 10\% the parameters of other models. Compared to fine-tuning GPT-4o with only MIMIC-CXR data for the same task, MedImageInsight outperforms in clinical metrics, but underperforms on lexical metrics where GPT-4o sets a new SOTA. Importantly for regulatory purposes, MedImageInsight can generate ROC curves, adjust sensitivity and specificity based on clinical need, and provide evidence-based decision support through image-image search (which can also enable retrieval augmented generation). In an independent clinical evaluation of image-image search in chest X-ray, MedImageInsight outperformed every other publicly available foundation model evaluated by large margins (over 6 points AUC), and significantly outperformed other models in terms of AI fairness (across age and gender). We hope releasing MedImageInsight will help enhance collective progress in medical imaging AI research and development.
3D TransUNet: Advancing Medical Image Segmentation through Vision Transformers
Oct 11, 2023Abstract:Medical image segmentation plays a crucial role in advancing healthcare systems for disease diagnosis and treatment planning. The u-shaped architecture, popularly known as U-Net, has proven highly successful for various medical image segmentation tasks. However, U-Net's convolution-based operations inherently limit its ability to model long-range dependencies effectively. To address these limitations, researchers have turned to Transformers, renowned for their global self-attention mechanisms, as alternative architectures. One popular network is our previous TransUNet, which leverages Transformers' self-attention to complement U-Net's localized information with the global context. In this paper, we extend the 2D TransUNet architecture to a 3D network by building upon the state-of-the-art nnU-Net architecture, and fully exploring Transformers' potential in both the encoder and decoder design. We introduce two key components: 1) A Transformer encoder that tokenizes image patches from a convolution neural network (CNN) feature map, enabling the extraction of global contexts, and 2) A Transformer decoder that adaptively refines candidate regions by utilizing cross-attention between candidate proposals and U-Net features. Our investigations reveal that different medical tasks benefit from distinct architectural designs. The Transformer encoder excels in multi-organ segmentation, where the relationship among organs is crucial. On the other hand, the Transformer decoder proves more beneficial for dealing with small and challenging segmented targets such as tumor segmentation. Extensive experiments showcase the significant potential of integrating a Transformer-based encoder and decoder into the u-shaped medical image segmentation architecture. TransUNet outperforms competitors in various medical applications.
The Effect of Counterfactuals on Reading Chest X-rays
Apr 02, 2023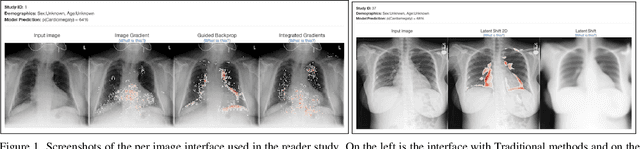
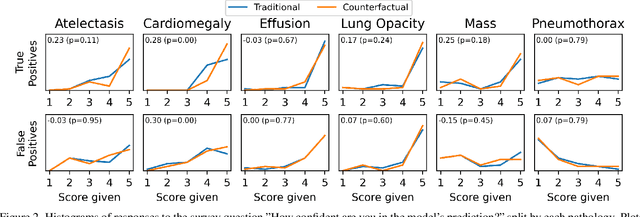
Abstract:This study evaluates the effect of counterfactual explanations on the interpretation of chest X-rays. We conduct a reader study with two radiologists assessing 240 chest X-ray predictions to rate their confidence that the model's prediction is correct using a 5 point scale. Half of the predictions are false positives. Each prediction is explained twice, once using traditional attribution methods and once with a counterfactual explanation. The overall results indicate that counterfactual explanations allow a radiologist to have more confidence in true positive predictions compared to traditional approaches (0.15$\pm$0.95 with p=0.01) with only a small increase in false positive predictions (0.04$\pm$1.06 with p=0.57). We observe the specific prediction tasks of Mass and Atelectasis appear to benefit the most compared to other tasks.
Current State of Community-Driven Radiological AI Deployment in Medical Imaging
Dec 29, 2022



Abstract:Artificial Intelligence (AI) has become commonplace to solve routine everyday tasks. Because of the exponential growth in medical imaging data volume and complexity, the workload on radiologists is steadily increasing. We project that the gap between the number of imaging exams and the number of expert radiologist readers required to cover this increase will continue to expand, consequently introducing a demand for AI-based tools that improve the efficiency with which radiologists can comfortably interpret these exams. AI has been shown to improve efficiency in medical-image generation, processing, and interpretation, and a variety of such AI models have been developed across research labs worldwide. However, very few of these, if any, find their way into routine clinical use, a discrepancy that reflects the divide between AI research and successful AI translation. To address the barrier to clinical deployment, we have formed MONAI Consortium, an open-source community which is building standards for AI deployment in healthcare institutions, and developing tools and infrastructure to facilitate their implementation. This report represents several years of weekly discussions and hands-on problem solving experience by groups of industry experts and clinicians in the MONAI Consortium. We identify barriers between AI-model development in research labs and subsequent clinical deployment and propose solutions. Our report provides guidance on processes which take an imaging AI model from development to clinical implementation in a healthcare institution. We discuss various AI integration points in a clinical Radiology workflow. We also present a taxonomy of Radiology AI use-cases. Through this report, we intend to educate the stakeholders in healthcare and AI (AI researchers, radiologists, imaging informaticists, and regulators) about cross-disciplinary challenges and possible solutions.
Who Goes First? Influences of Human-AI Workflow on Decision Making in Clinical Imaging
May 19, 2022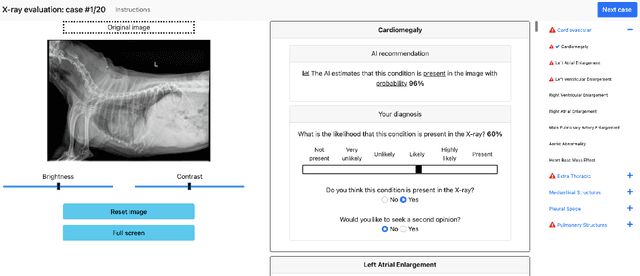
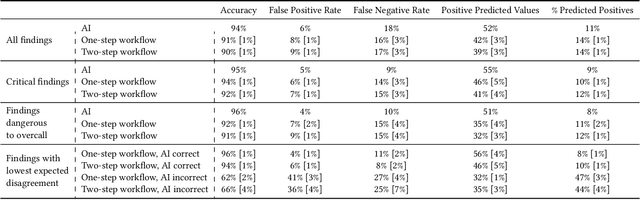
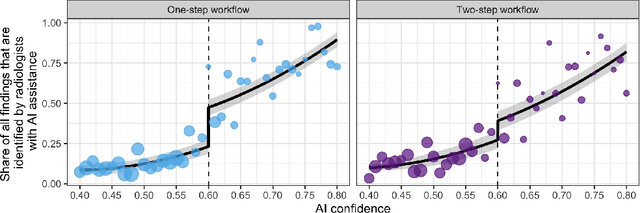
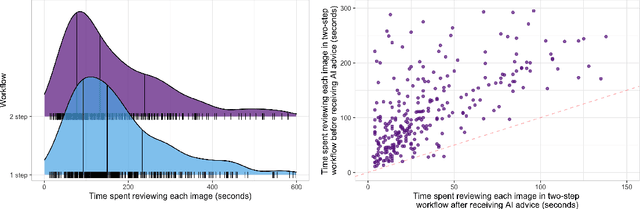
Abstract:Details of the designs and mechanisms in support of human-AI collaboration must be considered in the real-world fielding of AI technologies. A critical aspect of interaction design for AI-assisted human decision making are policies about the display and sequencing of AI inferences within larger decision-making workflows. We have a poor understanding of the influences of making AI inferences available before versus after human review of a diagnostic task at hand. We explore the effects of providing AI assistance at the start of a diagnostic session in radiology versus after the radiologist has made a provisional decision. We conducted a user study where 19 veterinary radiologists identified radiographic findings present in patients' X-ray images, with the aid of an AI tool. We employed two workflow configurations to analyze (i) anchoring effects, (ii) human-AI team diagnostic performance and agreement, (iii) time spent and confidence in decision making, and (iv) perceived usefulness of the AI. We found that participants who are asked to register provisional responses in advance of reviewing AI inferences are less likely to agree with the AI regardless of whether the advice is accurate and, in instances of disagreement with the AI, are less likely to seek the second opinion of a colleague. These participants also reported the AI advice to be less useful. Surprisingly, requiring provisional decisions on cases in advance of the display of AI inferences did not lengthen the time participants spent on the task. The study provides generalizable and actionable insights for the deployment of clinical AI tools in human-in-the-loop systems and introduces a methodology for studying alternative designs for human-AI collaboration. We make our experimental platform available as open source to facilitate future research on the influence of alternate designs on human-AI workflows.
Active label cleaning: Improving dataset quality under resource constraints
Sep 01, 2021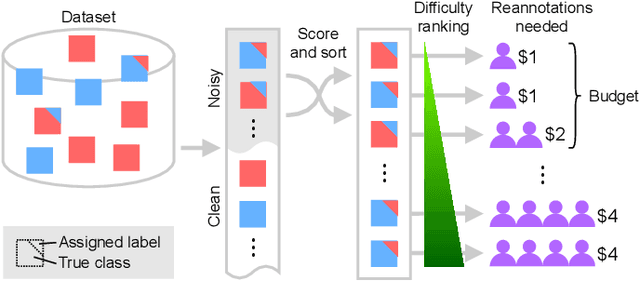
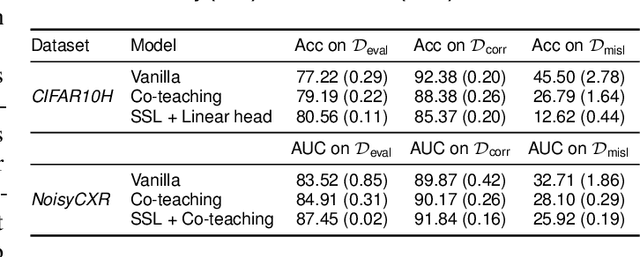
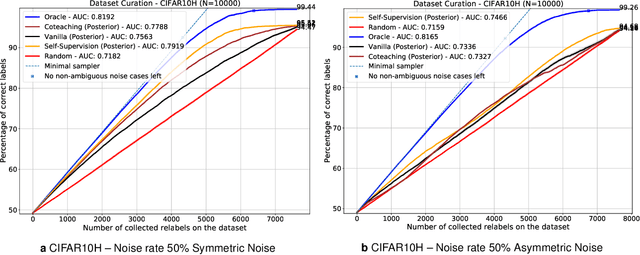
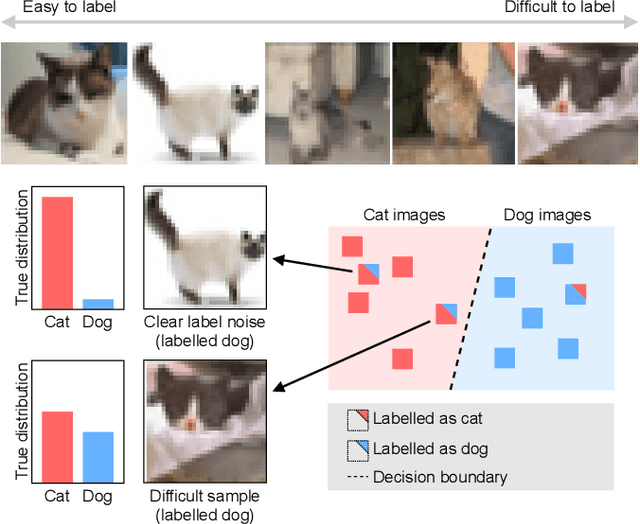
Abstract:Imperfections in data annotation, known as label noise, are detrimental to the training of machine learning models and have an often-overlooked confounding effect on the assessment of model performance. Nevertheless, employing experts to remove label noise by fully re-annotating large datasets is infeasible in resource-constrained settings, such as healthcare. This work advocates for a data-driven approach to prioritising samples for re-annotation - which we term "active label cleaning". We propose to rank instances according to estimated label correctness and labelling difficulty of each sample, and introduce a simulation framework to evaluate relabelling efficacy. Our experiments on natural images and on a new medical imaging benchmark show that cleaning noisy labels mitigates their negative impact on model training, evaluation, and selection. Crucially, the proposed active label cleaning enables correcting labels up to 4 times more effectively than typical random selection in realistic conditions, making better use of experts' valuable time for improving dataset quality.
CheXphotogenic: Generalization of Deep Learning Models for Chest X-ray Interpretation to Photos of Chest X-rays
Nov 12, 2020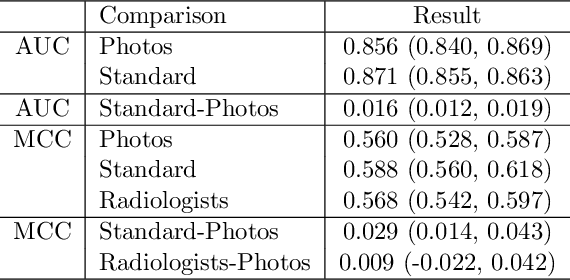
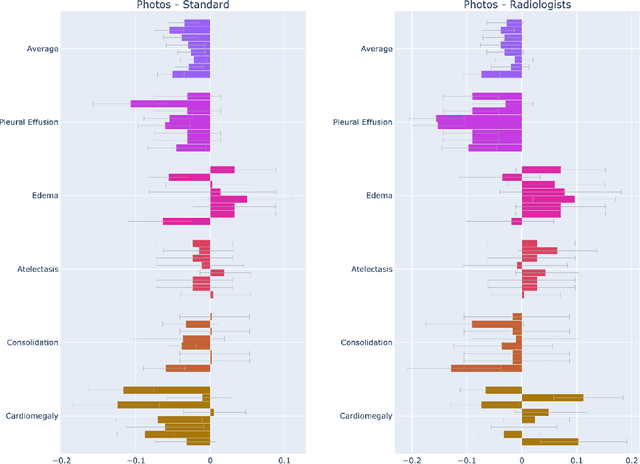
Abstract:The use of smartphones to take photographs of chest x-rays represents an appealing solution for scaled deployment of deep learning models for chest x-ray interpretation. However, the performance of chest x-ray algorithms on photos of chest x-rays has not been thoroughly investigated. In this study, we measured the diagnostic performance for 8 different chest x-ray models when applied to photos of chest x-rays. All models were developed by different groups and submitted to the CheXpert challenge, and re-applied to smartphone photos of x-rays in the CheXphoto dataset without further tuning. We found that several models had a drop in performance when applied to photos of chest x-rays, but even with this drop, some models still performed comparably to radiologists. Further investigation could be directed towards understanding how different model training procedures may affect model generalization to photos of chest x-rays.
Cross-Modal Data Programming Enables Rapid Medical Machine Learning
Mar 26, 2019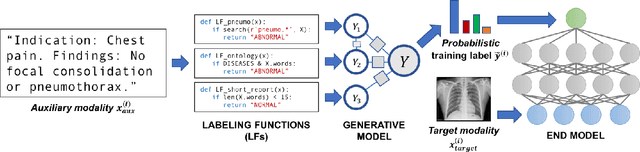
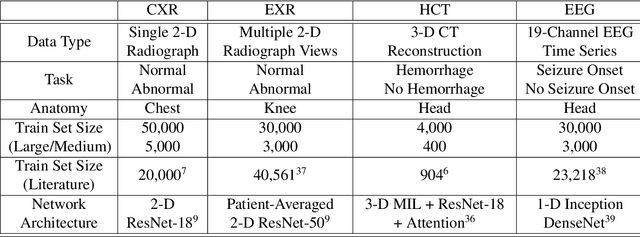
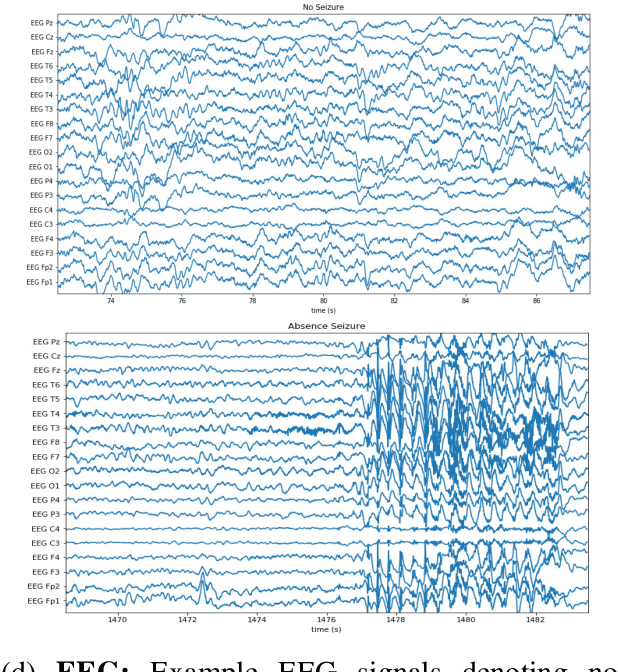
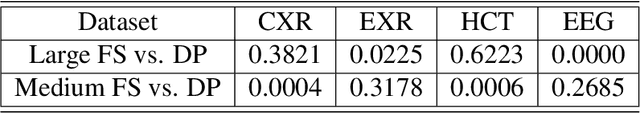
Abstract:Labeling training datasets has become a key barrier to building medical machine learning models. One strategy is to generate training labels programmatically, for example by applying natural language processing pipelines to text reports associated with imaging studies. We propose cross-modal data programming, which generalizes this intuitive strategy in a theoretically-grounded way that enables simpler, clinician-driven input, reduces required labeling time, and improves with additional unlabeled data. In this approach, clinicians generate training labels for models defined over a target modality (e.g. images or time series) by writing rules over an auxiliary modality (e.g. text reports). The resulting technical challenge consists of estimating the accuracies and correlations of these rules; we extend a recent unsupervised generative modeling technique to handle this cross-modal setting in a provably consistent way. Across four applications in radiography, computed tomography, and electroencephalography, and using only several hours of clinician time, our approach matches or exceeds the efficacy of physician-months of hand-labeling with statistical significance, demonstrating a fundamentally faster and more flexible way of building machine learning models in medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge