Melanie Bernhardt
Risk of Bias in Chest X-ray Foundation Models
Sep 07, 2022

Abstract:Foundation models are considered a breakthrough in all applications of AI, promising robust and reusable mechanisms for feature extraction, alleviating the need for large amounts of high quality training data for task-specific prediction models. However, foundation models may potentially encode and even reinforce existing biases present in historic datasets. Given the limited ability to scrutinize foundation models, it remains unclear whether the opportunities outweigh the risks in safety critical applications such as clinical decision making. In our statistical bias analysis of a recently published, and publicly available chest X-ray foundation model, we found reasons for concern as the model seems to encode protected characteristics including biological sex and racial identity, which may lead to disparate performance across subgroups in downstream applications. While research into foundation models for healthcare applications is in an early stage, we believe it is important to make the community aware of these risks to avoid harm.
Failure Detection in Medical Image Classification: A Reality Check and Benchmarking Testbed
May 27, 2022



Abstract:Failure detection in automated image classification is a critical safeguard for clinical deployment. Detected failure cases can be referred to human assessment, ensuring patient safety in computer-aided clinical decision making. Despite its paramount importance, there is insufficient evidence about the ability of state-of-the-art confidence scoring methods to detect test-time failures of classification models in the context of medical imaging. This paper provides a reality check, establishing the performance of in-domain misclassification detection methods, benchmarking 9 confidence scores on 6 medical imaging datasets with different imaging modalities, in multiclass and binary classification settings. Our experiments show that the problem of failure detection is far from being solved. We found that none of the benchmarked advanced methods proposed in the computer vision and machine learning literature can consistently outperform a simple softmax baseline. Our developed testbed facilitates future work in this important area.
Investigating underdiagnosis of AI algorithms in the presence of multiple sources of dataset bias
Jan 19, 2022Abstract:Deep learning models have shown great potential for image-based diagnosis assisting clinical decision making. At the same time, an increasing number of reports raise concerns about the potential risk that machine learning could amplify existing health disparities due to human biases that are embedded in the training data. It is of great importance to carefully investigate the extent to which biases may be reproduced or even amplified if we wish to build fair artificial intelligence systems. Seyyed-Kalantari et al. advance this conversation by analysing the performance of a disease classifier across population subgroups. They raise performance disparities related to underdiagnosis as a point of concern; we identify areas from this analysis which we believe deserve additional attention. Specifically, we wish to highlight some theoretical and practical difficulties associated with assessing model fairness through testing on data drawn from the same biased distribution as the training data, especially when the sources and amount of biases are unknown.
Active label cleaning: Improving dataset quality under resource constraints
Sep 01, 2021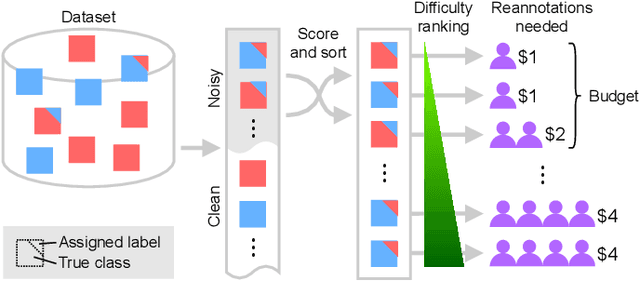
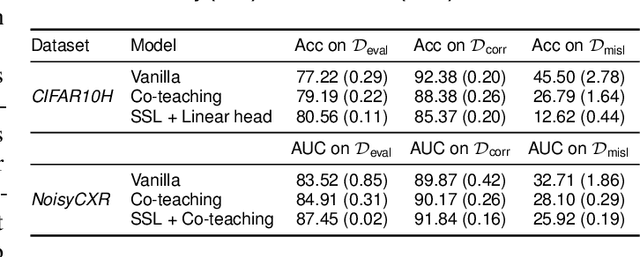
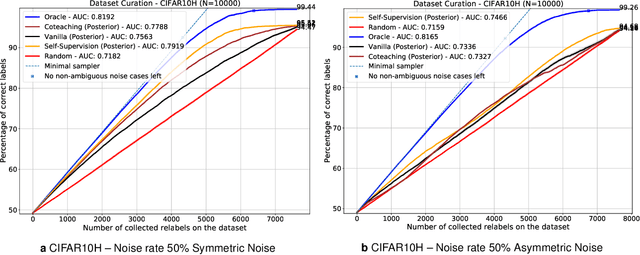
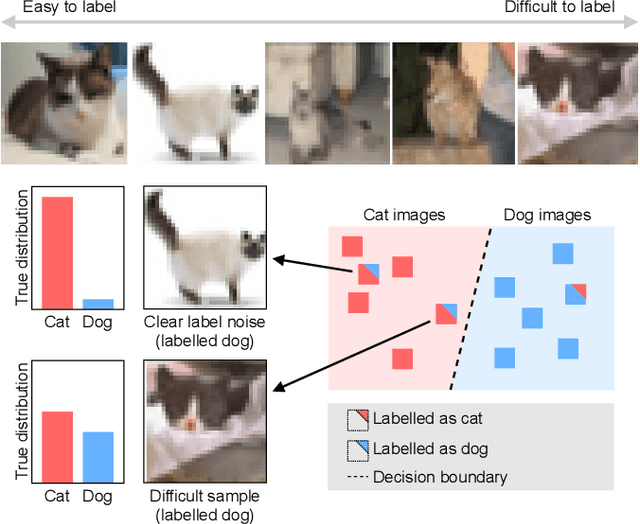
Abstract:Imperfections in data annotation, known as label noise, are detrimental to the training of machine learning models and have an often-overlooked confounding effect on the assessment of model performance. Nevertheless, employing experts to remove label noise by fully re-annotating large datasets is infeasible in resource-constrained settings, such as healthcare. This work advocates for a data-driven approach to prioritising samples for re-annotation - which we term "active label cleaning". We propose to rank instances according to estimated label correctness and labelling difficulty of each sample, and introduce a simulation framework to evaluate relabelling efficacy. Our experiments on natural images and on a new medical imaging benchmark show that cleaning noisy labels mitigates their negative impact on model training, evaluation, and selection. Crucially, the proposed active label cleaning enables correcting labels up to 4 times more effectively than typical random selection in realistic conditions, making better use of experts' valuable time for improving dataset quality.
Hierarchical Analysis of Visual COVID-19 Features from Chest Radiographs
Jul 14, 2021

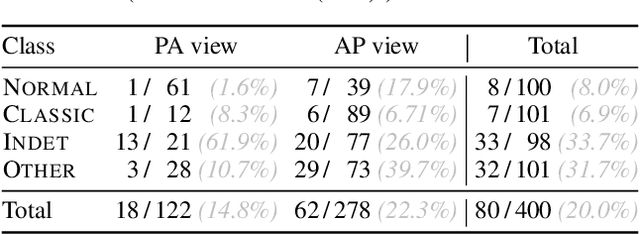

Abstract:Chest radiography has been a recommended procedure for patient triaging and resource management in intensive care units (ICUs) throughout the COVID-19 pandemic. The machine learning efforts to augment this workflow have been long challenged due to deficiencies in reporting, model evaluation, and failure mode analysis. To address some of those shortcomings, we model radiological features with a human-interpretable class hierarchy that aligns with the radiological decision process. Also, we propose the use of a data-driven error analysis methodology to uncover the blind spots of our model, providing further transparency on its clinical utility. For example, our experiments show that model failures highly correlate with ICU imaging conditions and with the inherent difficulty in distinguishing certain types of radiological features. Also, our hierarchical interpretation and analysis facilitates the comparison with respect to radiologists' findings and inter-variability, which in return helps us to better assess the clinical applicability of models.
Training Variational Networks with Multi-Domain Simulations: Speed-of-Sound Image Reconstruction
Jun 25, 2020



Abstract:Speed-of-sound has been shown as a potential biomarker for breast cancer imaging, successfully differentiating malignant tumors from benign ones. Speed-of-sound images can be reconstructed from time-of-flight measurements from ultrasound images acquired using conventional handheld ultrasound transducers. Variational Networks (VN) have recently been shown to be a potential learning-based approach for optimizing inverse problems in image reconstruction. Despite earlier promising results, these methods however do not generalize well from simulated to acquired data, due to the domain shift. In this work, we present for the first time a VN solution for a pulse-echo SoS image reconstruction problem using diverging waves with conventional transducers and single-sided tissue access. This is made possible by incorporating simulations with varying complexity into training. We use loop unrolling of gradient descent with momentum, with an exponentially weighted loss of outputs at each unrolled iteration in order to regularize training. We learn norms as activation functions regularized to have smooth forms for robustness to input distribution variations. We evaluate reconstruction quality on ray-based and full-wave simulations as well as on tissue-mimicking phantom data, in comparison to a classical iterative (L-BFGS) optimization of this image reconstruction problem. We show that the proposed regularization techniques combined with multi-source domain training yield substantial improvements in the domain adaptation capabilities of VN, reducing median RMSE by 54% on a wave-based simulation dataset compared to the baseline VN. We also show that on data acquired from a tissue-mimicking breast phantom the proposed VN provides improved reconstruction in 12 milliseconds.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge