Miguel Monteiro
High Fidelity Image Counterfactuals with Probabilistic Causal Models
Jul 18, 2023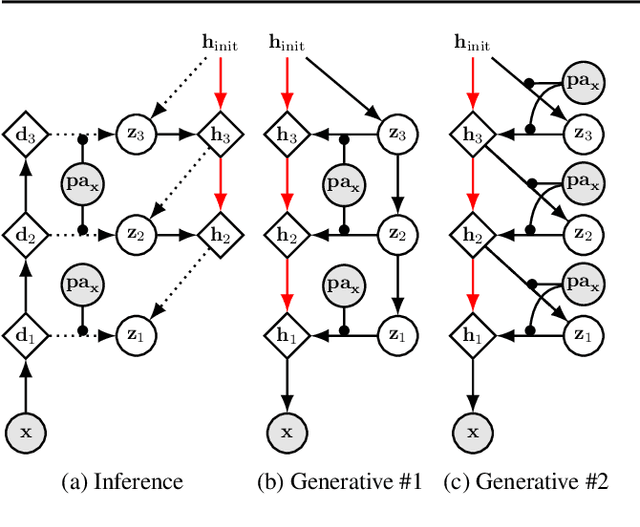
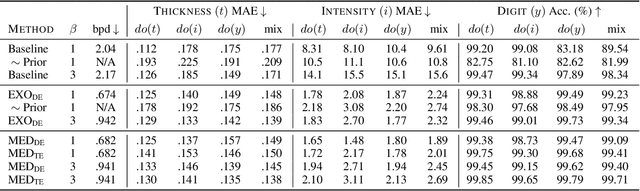
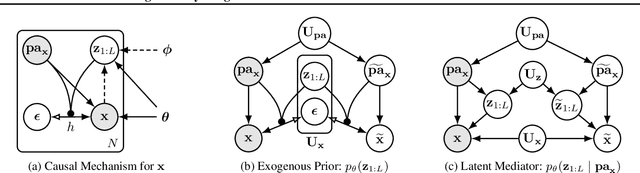
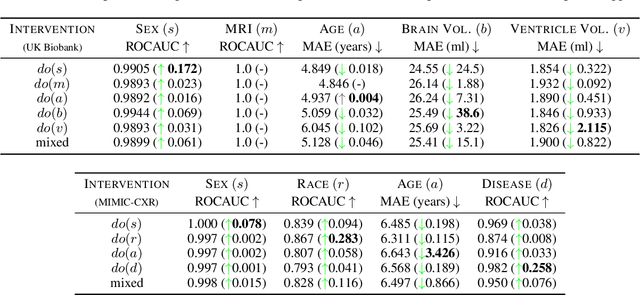
Abstract:We present a general causal generative modelling framework for accurate estimation of high fidelity image counterfactuals with deep structural causal models. Estimation of interventional and counterfactual queries for high-dimensional structured variables, such as images, remains a challenging task. We leverage ideas from causal mediation analysis and advances in generative modelling to design new deep causal mechanisms for structured variables in causal models. Our experiments demonstrate that our proposed mechanisms are capable of accurate abduction and estimation of direct, indirect and total effects as measured by axiomatic soundness of counterfactuals.
Measuring axiomatic soundness of counterfactual image models
Mar 02, 2023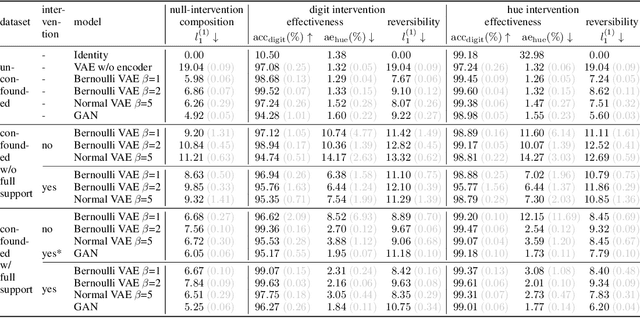
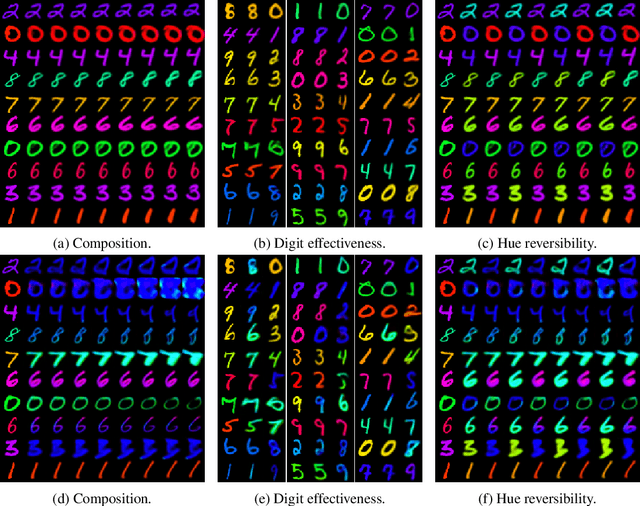
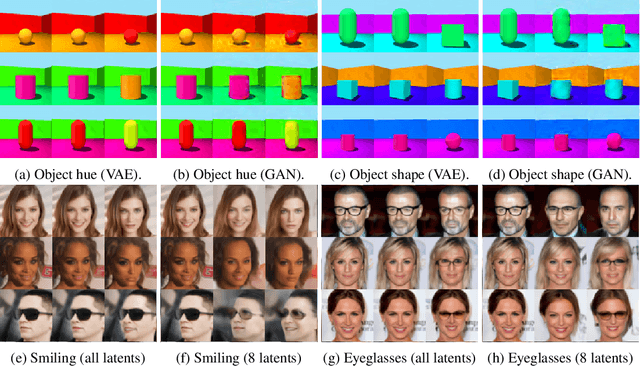
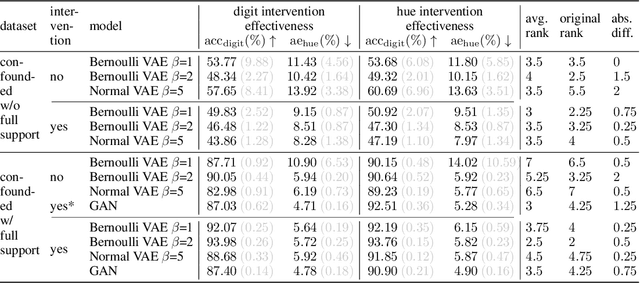
Abstract:We present a general framework for evaluating image counterfactuals. The power and flexibility of deep generative models make them valuable tools for learning mechanisms in structural causal models. However, their flexibility makes counterfactual identifiability impossible in the general case. Motivated by these issues, we revisit Pearl's axiomatic definition of counterfactuals to determine the necessary constraints of any counterfactual inference model: composition, reversibility, and effectiveness. We frame counterfactuals as functions of an input variable, its parents, and counterfactual parents and use the axiomatic constraints to restrict the set of functions that could represent the counterfactual, thus deriving distance metrics between the approximate and ideal functions. We demonstrate how these metrics can be used to compare and choose between different approximate counterfactual inference models and to provide insight into a model's shortcomings and trade-offs.
* Counterfactual inference, Generative Models, Computer Vision, Published in ICLR 2023
Analysing the effectiveness of a generative model for semi-supervised medical image segmentation
Nov 03, 2022



Abstract:Image segmentation is important in medical imaging, providing valuable, quantitative information for clinical decision-making in diagnosis, therapy, and intervention. The state-of-the-art in automated segmentation remains supervised learning, employing discriminative models such as U-Net. However, training these models requires access to large amounts of manually labelled data which is often difficult to obtain in real medical applications. In such settings, semi-supervised learning (SSL) attempts to leverage the abundance of unlabelled data to obtain more robust and reliable models. Recently, generative models have been proposed for semantic segmentation, as they make an attractive choice for SSL. Their ability to capture the joint distribution over input images and output label maps provides a natural way to incorporate information from unlabelled images. This paper analyses whether deep generative models such as the SemanticGAN are truly viable alternatives to tackle challenging medical image segmentation problems. To that end, we thoroughly evaluate the segmentation performance, robustness, and potential subgroup disparities of discriminative and generative segmentation methods when applied to large-scale, publicly available chest X-ray datasets.
Automatic lesion analysis for increased efficiency in outcome prediction of traumatic brain injury
Aug 08, 2022



Abstract:The accurate prognosis for traumatic brain injury (TBI) patients is difficult yet essential to inform therapy, patient management, and long-term after-care. Patient characteristics such as age, motor and pupil responsiveness, hypoxia and hypotension, and radiological findings on computed tomography (CT), have been identified as important variables for TBI outcome prediction. CT is the acute imaging modality of choice in clinical practice because of its acquisition speed and widespread availability. However, this modality is mainly used for qualitative and semi-quantitative assessment, such as the Marshall scoring system, which is prone to subjectivity and human errors. This work explores the predictive power of imaging biomarkers extracted from routinely-acquired hospital admission CT scans using a state-of-the-art, deep learning TBI lesion segmentation method. We use lesion volumes and corresponding lesion statistics as inputs for an extended TBI outcome prediction model. We compare the predictive power of our proposed features to the Marshall score, independently and when paired with classic TBI biomarkers. We find that automatically extracted quantitative CT features perform similarly or better than the Marshall score in predicting unfavourable TBI outcomes. Leveraging automatic atlas alignment, we also identify frontal extra-axial lesions as important indicators of poor outcome. Our work may contribute to a better understanding of TBI, and provides new insights into how automated neuroimaging analysis can be used to improve prognostication after TBI.
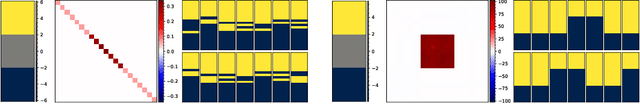
Structured Uncertainty in the Observation Space of Variational Autoencoders
May 25, 2022



Abstract:Variational autoencoders (VAEs) are a popular class of deep generative models with many variants and a wide range of applications. Improvements upon the standard VAE mostly focus on the modelling of the posterior distribution over the latent space and the properties of the neural network decoder. In contrast, improving the model for the observational distribution is rarely considered and typically defaults to a pixel-wise independent categorical or normal distribution. In image synthesis, sampling from such distributions produces spatially-incoherent results with uncorrelated pixel noise, resulting in only the sample mean being somewhat useful as an output prediction. In this paper, we aim to stay true to VAE theory by improving the samples from the observational distribution. We propose an alternative model for the observation space, encoding spatial dependencies via a low-rank parameterisation. We demonstrate that this new observational distribution has the ability to capture relevant covariance between pixels, resulting in spatially-coherent samples. In contrast to pixel-wise independent distributions, our samples seem to contain semantically meaningful variations from the mean allowing the prediction of multiple plausible outputs with a single forward pass.
Active label cleaning: Improving dataset quality under resource constraints
Sep 01, 2021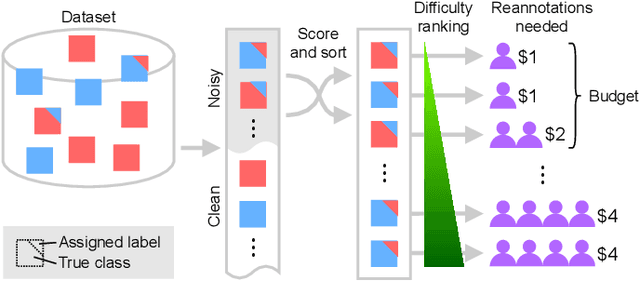
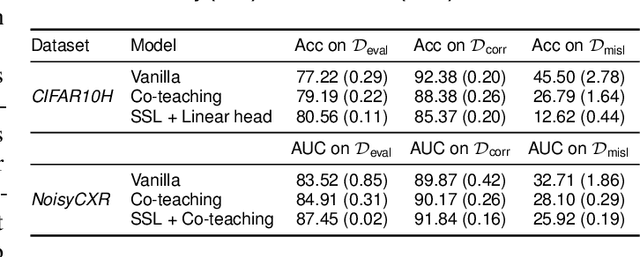
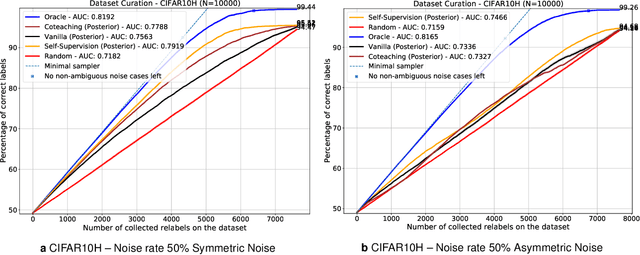
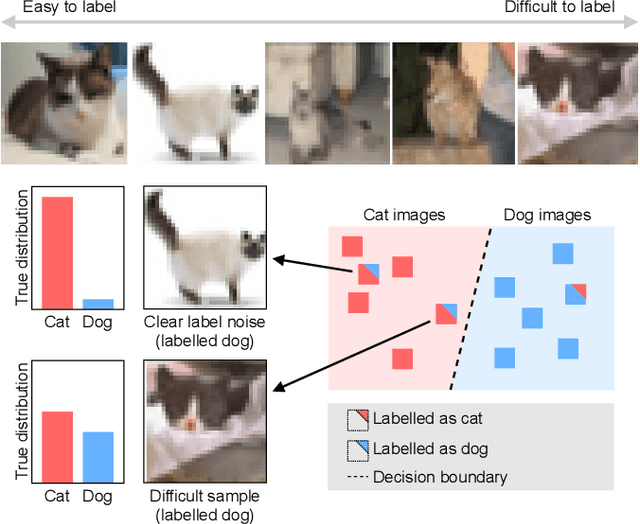
Abstract:Imperfections in data annotation, known as label noise, are detrimental to the training of machine learning models and have an often-overlooked confounding effect on the assessment of model performance. Nevertheless, employing experts to remove label noise by fully re-annotating large datasets is infeasible in resource-constrained settings, such as healthcare. This work advocates for a data-driven approach to prioritising samples for re-annotation - which we term "active label cleaning". We propose to rank instances according to estimated label correctness and labelling difficulty of each sample, and introduce a simulation framework to evaluate relabelling efficacy. Our experiments on natural images and on a new medical imaging benchmark show that cleaning noisy labels mitigates their negative impact on model training, evaluation, and selection. Crucially, the proposed active label cleaning enables correcting labels up to 4 times more effectively than typical random selection in realistic conditions, making better use of experts' valuable time for improving dataset quality.
Stochastic Segmentation Networks: Modelling Spatially Correlated Aleatoric Uncertainty
Jun 10, 2020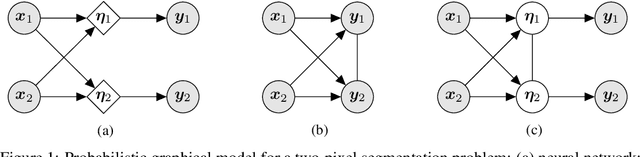

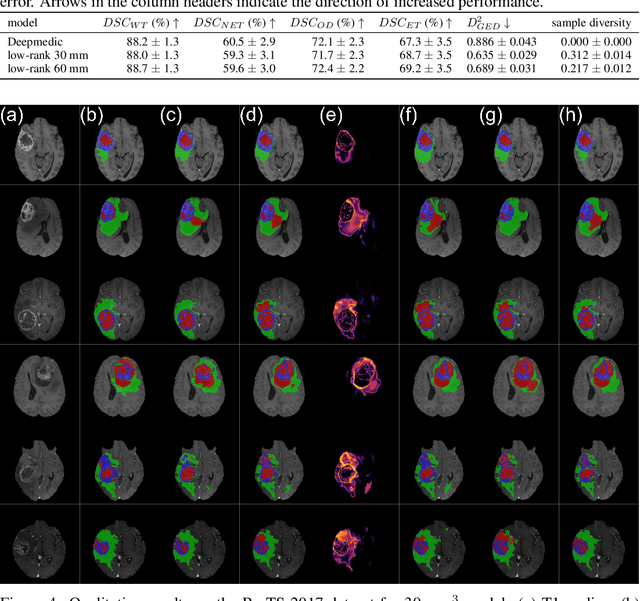
Abstract:In image segmentation, there is often more than one plausible solution for a given input. In medical imaging, for example, experts will often disagree about the exact location of object boundaries. Estimating this inherent uncertainty and predicting multiple plausible hypotheses is of great interest in many applications, yet this ability is lacking in most current deep learning methods. In this paper, we introduce stochastic segmentation networks (SSNs), an efficient probabilistic method for modelling aleatoric uncertainty with any image segmentation network architecture. In contrast to approaches that produce pixel-wise estimates, SSNs model joint distributions over entire label maps and thus can generate multiple spatially coherent hypotheses for a single image. By using a low-rank multivariate normal distribution over the logit space to model the probability of the label map given the image, we obtain a spatially consistent probability distribution that can be efficiently computed by a neural network without any changes to the underlying architecture. We tested our method on the segmentation of real-world medical data, including lung nodules in 2D CT and brain tumours in 3D multimodal MRI scans. SSNs outperform state-of-the-art for modelling correlated uncertainty in ambiguous images while being much simpler, more flexible, and more efficient.
Conditional Random Fields as Recurrent Neural Networks for 3D Medical Imaging Segmentation
Jul 19, 2018



Abstract:The Conditional Random Field as a Recurrent Neural Network layer is a recently proposed algorithm meant to be placed on top of an existing Fully-Convolutional Neural Network to improve the quality of semantic segmentation. In this paper, we test whether this algorithm, which was shown to improve semantic segmentation for 2D RGB images, is able to improve segmentation quality for 3D multi-modal medical images. We developed an implementation of the algorithm which works for any number of spatial dimensions, input/output image channels, and reference image channels. As far as we know this is the first publicly available implementation of this sort. We tested the algorithm with two distinct 3D medical imaging datasets, we concluded that the performance differences observed were not statistically significant. Finally, in the discussion section of the paper, we go into the reasons as to why this technique transfers poorly from natural images to medical images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge