Fabio De Sousa Ribeiro
Decoupled Classifier-Free Guidance for Counterfactual Diffusion Models
Jun 17, 2025Abstract:Counterfactual image generation aims to simulate realistic visual outcomes under specific causal interventions. Diffusion models have recently emerged as a powerful tool for this task, combining DDIM inversion with conditional generation via classifier-free guidance (CFG). However, standard CFG applies a single global weight across all conditioning variables, which can lead to poor identity preservation and spurious attribute changes - a phenomenon known as attribute amplification. To address this, we propose Decoupled Classifier-Free Guidance (DCFG), a flexible and model-agnostic framework that introduces group-wise conditioning control. DCFG builds on an attribute-split embedding strategy that disentangles semantic inputs, enabling selective guidance on user-defined attribute groups. For counterfactual generation, we partition attributes into intervened and invariant sets based on a causal graph and apply distinct guidance to each. Experiments on CelebA-HQ, MIMIC-CXR, and EMBED show that DCFG improves intervention fidelity, mitigates unintended changes, and enhances reversibility, enabling more faithful and interpretable counterfactual image generation.
Diffusion Counterfactual Generation with Semantic Abduction
Jun 09, 2025Abstract:Counterfactual image generation presents significant challenges, including preserving identity, maintaining perceptual quality, and ensuring faithfulness to an underlying causal model. While existing auto-encoding frameworks admit semantic latent spaces which can be manipulated for causal control, they struggle with scalability and fidelity. Advancements in diffusion models present opportunities for improving counterfactual image editing, having demonstrated state-of-the-art visual quality, human-aligned perception and representation learning capabilities. Here, we present a suite of diffusion-based causal mechanisms, introducing the notions of spatial, semantic and dynamic abduction. We propose a general framework that integrates semantic representations into diffusion models through the lens of Pearlian causality to edit images via a counterfactual reasoning process. To our knowledge, this is the first work to consider high-level semantic identity preservation for diffusion counterfactuals and to demonstrate how semantic control enables principled trade-offs between faithful causal control and identity preservation.
* Proceedings of the 42nd International Conference on Machine Learning, Vancouver, Canada
Robust image representations with counterfactual contrastive learning
Sep 16, 2024



Abstract:Contrastive pretraining can substantially increase model generalisation and downstream performance. However, the quality of the learned representations is highly dependent on the data augmentation strategy applied to generate positive pairs. Positive contrastive pairs should preserve semantic meaning while discarding unwanted variations related to the data acquisition domain. Traditional contrastive pipelines attempt to simulate domain shifts through pre-defined generic image transformations. However, these do not always mimic realistic and relevant domain variations for medical imaging such as scanner differences. To tackle this issue, we herein introduce counterfactual contrastive learning, a novel framework leveraging recent advances in causal image synthesis to create contrastive positive pairs that faithfully capture relevant domain variations. Our method, evaluated across five datasets encompassing both chest radiography and mammography data, for two established contrastive objectives (SimCLR and DINO-v2), outperforms standard contrastive learning in terms of robustness to acquisition shift. Notably, counterfactual contrastive learning achieves superior downstream performance on both in-distribution and on external datasets, especially for images acquired with scanners under-represented in the training set. Further experiments show that the proposed framework extends beyond acquisition shifts, with models trained with counterfactual contrastive learning substantially improving subgroup performance across biological sex.
Latent 3D Brain MRI Counterfactual
Sep 09, 2024



Abstract:The number of samples in structural brain MRI studies is often too small to properly train deep learning models. Generative models show promise in addressing this issue by effectively learning the data distribution and generating high-fidelity MRI. However, they struggle to produce diverse, high-quality data outside the distribution defined by the training data. One way to address the issue is using causal models developed for 3D volume counterfactuals. However, accurately modeling causality in high-dimensional spaces is a challenge so that these models generally generate 3D brain MRIS of lower quality. To address these challenges, we propose a two-stage method that constructs a Structural Causal Model (SCM) within the latent space. In the first stage, we employ a VQ-VAE to learn a compact embedding of the MRI volume. Subsequently, we integrate our causal model into this latent space and execute a three-step counterfactual procedure using a closed-form Generalized Linear Model (GLM). Our experiments conducted on real-world high-resolution MRI data (1mm) demonstrate that our method can generate high-quality 3D MRI counterfactuals.
Identifiable Object-Centric Representation Learning via Probabilistic Slot Attention
Jun 11, 2024
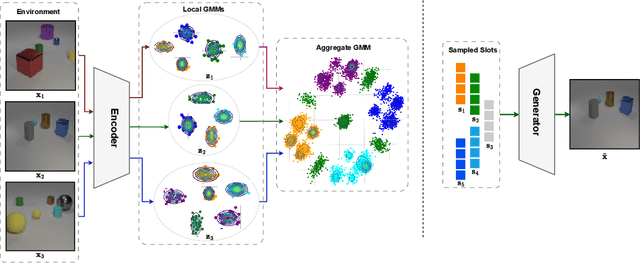
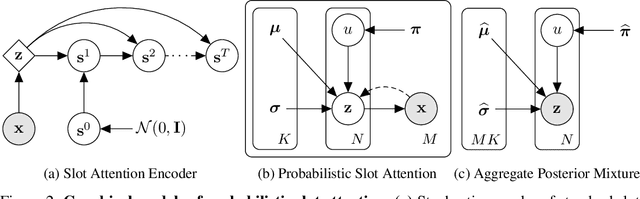
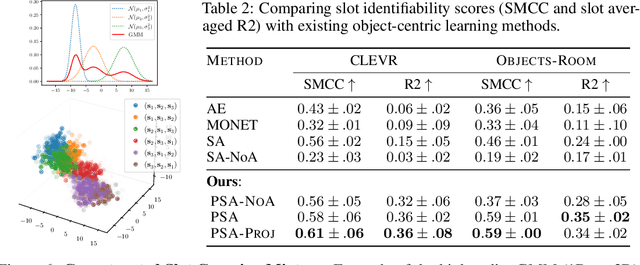
Abstract:Learning modular object-centric representations is crucial for systematic generalization. Existing methods show promising object-binding capabilities empirically, but theoretical identifiability guarantees remain relatively underdeveloped. Understanding when object-centric representations can theoretically be identified is crucial for scaling slot-based methods to high-dimensional images with correctness guarantees. To that end, we propose a probabilistic slot-attention algorithm that imposes an aggregate mixture prior over object-centric slot representations, thereby providing slot identifiability guarantees without supervision, up to an equivalence relation. We provide empirical verification of our theoretical identifiability result using both simple 2-dimensional data and high-resolution imaging datasets.
Mitigating attribute amplification in counterfactual image generation
Mar 14, 2024



Abstract:Causal generative modelling is gaining interest in medical imaging due to its ability to answer interventional and counterfactual queries. Most work focuses on generating counterfactual images that look plausible, using auxiliary classifiers to enforce effectiveness of simulated interventions. We investigate pitfalls in this approach, discovering the issue of attribute amplification, where unrelated attributes are spuriously affected during interventions, leading to biases across protected characteristics and disease status. We show that attribute amplification is caused by the use of hard labels in the counterfactual training process and propose soft counterfactual fine-tuning to mitigate this issue. Our method substantially reduces the amplification effect while maintaining effectiveness of generated images, demonstrated on a large chest X-ray dataset. Our work makes an important advancement towards more faithful and unbiased causal modelling in medical imaging.
Counterfactual contrastive learning: robust representations via causal image synthesis
Mar 14, 2024Abstract:Contrastive pretraining is well-known to improve downstream task performance and model generalisation, especially in limited label settings. However, it is sensitive to the choice of augmentation pipeline. Positive pairs should preserve semantic information while destroying domain-specific information. Standard augmentation pipelines emulate domain-specific changes with pre-defined photometric transformations, but what if we could simulate realistic domain changes instead? In this work, we show how to utilise recent progress in counterfactual image generation to this effect. We propose CF-SimCLR, a counterfactual contrastive learning approach which leverages approximate counterfactual inference for positive pair creation. Comprehensive evaluation across five datasets, on chest radiography and mammography, demonstrates that CF-SimCLR substantially improves robustness to acquisition shift with higher downstream performance on both in- and out-of-distribution data, particularly for domains which are under-represented during training.
Demystifying Variational Diffusion Models
Jan 11, 2024



Abstract:Despite the growing popularity of diffusion models, gaining a deep understanding of the model class remains somewhat elusive for the uninitiated in non-equilibrium statistical physics. With that in mind, we present what we believe is a more straightforward introduction to diffusion models using directed graphical modelling and variational Bayesian principles, which imposes relatively fewer prerequisites on the average reader. Our exposition constitutes a comprehensive technical review spanning from foundational concepts like deep latent variable models to recent advances in continuous-time diffusion-based modelling, highlighting theoretical connections between model classes along the way. We provide additional mathematical insights that were omitted in the seminal works whenever possible to aid in understanding, while avoiding the introduction of new notation. We envision this article serving as a useful educational supplement for both researchers and practitioners in the area, and we welcome feedback and contributions from the community at https://github.com/biomedia-mira/demystifying-diffusion.
No Fair Lunch: A Causal Perspective on Dataset Bias in Machine Learning for Medical Imaging
Jul 31, 2023



Abstract:As machine learning methods gain prominence within clinical decision-making, addressing fairness concerns becomes increasingly urgent. Despite considerable work dedicated to detecting and ameliorating algorithmic bias, today's methods are deficient with potentially harmful consequences. Our causal perspective sheds new light on algorithmic bias, highlighting how different sources of dataset bias may appear indistinguishable yet require substantially different mitigation strategies. We introduce three families of causal bias mechanisms stemming from disparities in prevalence, presentation, and annotation. Our causal analysis underscores how current mitigation methods tackle only a narrow and often unrealistic subset of scenarios. We provide a practical three-step framework for reasoning about fairness in medical imaging, supporting the development of safe and equitable AI prediction models.
High Fidelity Image Counterfactuals with Probabilistic Causal Models
Jul 18, 2023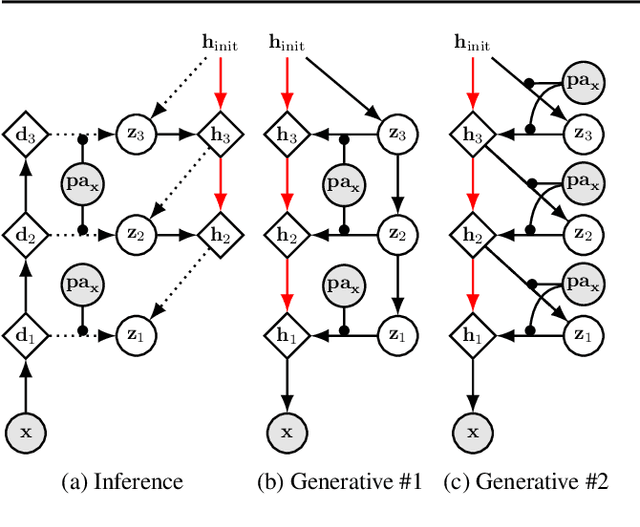
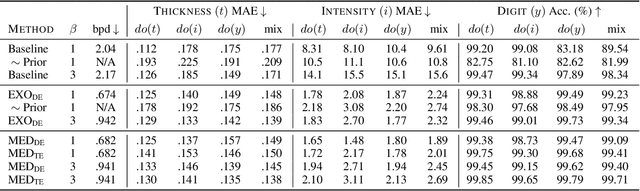
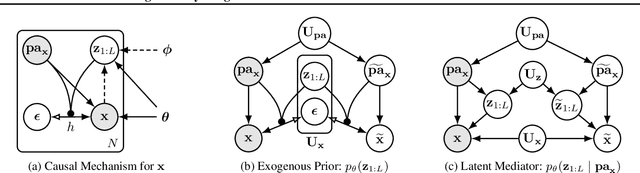
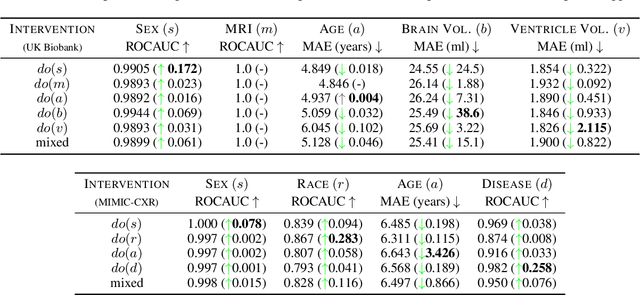
Abstract:We present a general causal generative modelling framework for accurate estimation of high fidelity image counterfactuals with deep structural causal models. Estimation of interventional and counterfactual queries for high-dimensional structured variables, such as images, remains a challenging task. We leverage ideas from causal mediation analysis and advances in generative modelling to design new deep causal mechanisms for structured variables in causal models. Our experiments demonstrate that our proposed mechanisms are capable of accurate abduction and estimation of direct, indirect and total effects as measured by axiomatic soundness of counterfactuals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge