Ben Glocker
Biomedical Image Analysis Group, Department of Computing, Imperial College London
A Primer on Causal and Statistical Dataset Biases for Fair and Robust Image Analysis
Sep 04, 2025Abstract:Machine learning methods often fail when deployed in the real world. Worse still, they fail in high-stakes situations and across socially sensitive lines. These issues have a chilling effect on the adoption of machine learning methods in settings such as medical diagnosis, where they are arguably best-placed to provide benefits if safely deployed. In this primer, we introduce the causal and statistical structures which induce failure in machine learning methods for image analysis. We highlight two previously overlooked problems, which we call the \textit{no fair lunch} problem and the \textit{subgroup separability} problem. We elucidate why today's fair representation learning methods fail to adequately solve them and propose potential paths forward for the field.
Exploring the interplay of label bias with subgroup size and separability: A case study in mammographic density classification
Jul 24, 2025Abstract:Systematic mislabelling affecting specific subgroups (i.e., label bias) in medical imaging datasets represents an understudied issue concerning the fairness of medical AI systems. In this work, we investigated how size and separability of subgroups affected by label bias influence the learned features and performance of a deep learning model. Therefore, we trained deep learning models for binary tissue density classification using the EMory BrEast imaging Dataset (EMBED), where label bias affected separable subgroups (based on imaging manufacturer) or non-separable "pseudo-subgroups". We found that simulated subgroup label bias led to prominent shifts in the learned feature representations of the models. Importantly, these shifts within the feature space were dependent on both the relative size and the separability of the subgroup affected by label bias. We also observed notable differences in subgroup performance depending on whether a validation set with clean labels was used to define the classification threshold for the model. For instance, with label bias affecting the majority separable subgroup, the true positive rate for that subgroup fell from 0.898, when the validation set had clean labels, to 0.518, when the validation set had biased labels. Our work represents a key contribution toward understanding the consequences of label bias on subgroup fairness in medical imaging AI.
Where are we with calibration under dataset shift in image classification?
Jul 10, 2025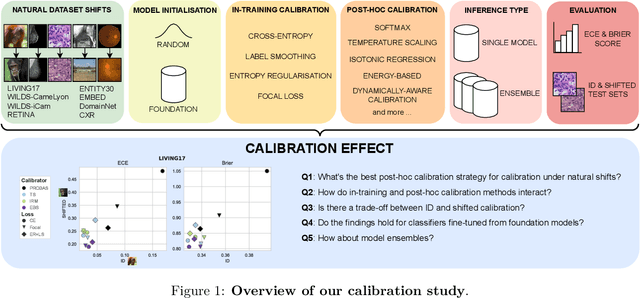
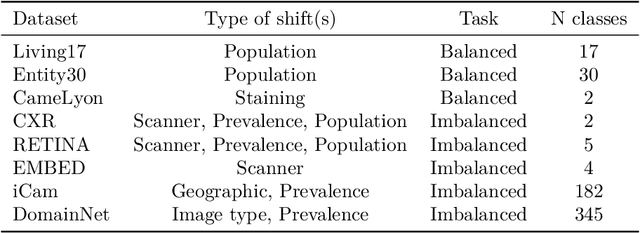
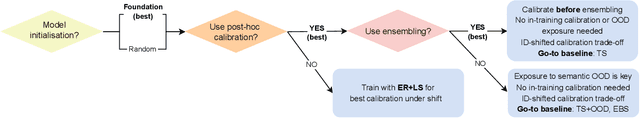
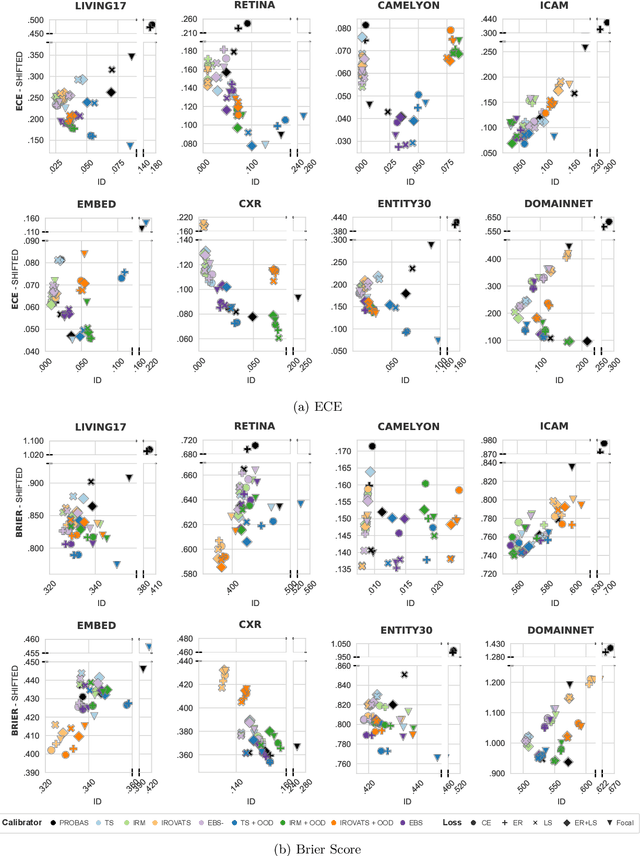
Abstract:We conduct an extensive study on the state of calibration under real-world dataset shift for image classification. Our work provides important insights on the choice of post-hoc and in-training calibration techniques, and yields practical guidelines for all practitioners interested in robust calibration under shift. We compare various post-hoc calibration methods, and their interactions with common in-training calibration strategies (e.g., label smoothing), across a wide range of natural shifts, on eight different classification tasks across several imaging domains. We find that: (i) simultaneously applying entropy regularisation and label smoothing yield the best calibrated raw probabilities under dataset shift, (ii) post-hoc calibrators exposed to a small amount of semantic out-of-distribution data (unrelated to the task) are most robust under shift, (iii) recent calibration methods specifically aimed at increasing calibration under shifts do not necessarily offer significant improvements over simpler post-hoc calibration methods, (iv) improving calibration under shifts often comes at the cost of worsening in-distribution calibration. Importantly, these findings hold for randomly initialised classifiers, as well as for those finetuned from foundation models, the latter being consistently better calibrated compared to models trained from scratch. Finally, we conduct an in-depth analysis of ensembling effects, finding that (i) applying calibration prior to ensembling (instead of after) is more effective for calibration under shifts, (ii) for ensembles, OOD exposure deteriorates the ID-shifted calibration trade-off, (iii) ensembling remains one of the most effective methods to improve calibration robustness and, combined with finetuning from foundation models, yields best calibration results overall.
Decoupled Classifier-Free Guidance for Counterfactual Diffusion Models
Jun 17, 2025Abstract:Counterfactual image generation aims to simulate realistic visual outcomes under specific causal interventions. Diffusion models have recently emerged as a powerful tool for this task, combining DDIM inversion with conditional generation via classifier-free guidance (CFG). However, standard CFG applies a single global weight across all conditioning variables, which can lead to poor identity preservation and spurious attribute changes - a phenomenon known as attribute amplification. To address this, we propose Decoupled Classifier-Free Guidance (DCFG), a flexible and model-agnostic framework that introduces group-wise conditioning control. DCFG builds on an attribute-split embedding strategy that disentangles semantic inputs, enabling selective guidance on user-defined attribute groups. For counterfactual generation, we partition attributes into intervened and invariant sets based on a causal graph and apply distinct guidance to each. Experiments on CelebA-HQ, MIMIC-CXR, and EMBED show that DCFG improves intervention fidelity, mitigates unintended changes, and enhances reversibility, enabling more faithful and interpretable counterfactual image generation.
Object-Centric Neuro-Argumentative Learning
Jun 17, 2025Abstract:Over the last decade, as we rely more on deep learning technologies to make critical decisions, concerns regarding their safety, reliability and interpretability have emerged. We introduce a novel Neural Argumentative Learning (NAL) architecture that integrates Assumption-Based Argumentation (ABA) with deep learning for image analysis. Our architecture consists of neural and symbolic components. The former segments and encodes images into facts using object-centric learning, while the latter applies ABA learning to develop ABA frameworks enabling predictions with images. Experiments on synthetic data show that the NAL architecture can be competitive with a state-of-the-art alternative.
Vector Representations of Vessel Trees
Jun 11, 2025Abstract:We introduce a novel framework for learning vector representations of tree-structured geometric data focusing on 3D vascular networks. Our approach employs two sequentially trained Transformer-based autoencoders. In the first stage, the Vessel Autoencoder captures continuous geometric details of individual vessel segments by learning embeddings from sampled points along each curve. In the second stage, the Vessel Tree Autoencoder encodes the topology of the vascular network as a single vector representation, leveraging the segment-level embeddings from the first model. A recursive decoding process ensures that the reconstructed topology is a valid tree structure. Compared to 3D convolutional models, this proposed approach substantially lowers GPU memory requirements, facilitating large-scale training. Experimental results on a 2D synthetic tree dataset and a 3D coronary artery dataset demonstrate superior reconstruction fidelity, accurate topology preservation, and realistic interpolations in latent space. Our scalable framework, named VeTTA, offers precise, flexible, and topologically consistent modeling of anatomical tree structures in medical imaging.
Diffusion Counterfactual Generation with Semantic Abduction
Jun 09, 2025Abstract:Counterfactual image generation presents significant challenges, including preserving identity, maintaining perceptual quality, and ensuring faithfulness to an underlying causal model. While existing auto-encoding frameworks admit semantic latent spaces which can be manipulated for causal control, they struggle with scalability and fidelity. Advancements in diffusion models present opportunities for improving counterfactual image editing, having demonstrated state-of-the-art visual quality, human-aligned perception and representation learning capabilities. Here, we present a suite of diffusion-based causal mechanisms, introducing the notions of spatial, semantic and dynamic abduction. We propose a general framework that integrates semantic representations into diffusion models through the lens of Pearlian causality to edit images via a counterfactual reasoning process. To our knowledge, this is the first work to consider high-level semantic identity preservation for diffusion counterfactuals and to demonstrate how semantic control enables principled trade-offs between faithful causal control and identity preservation.
* Proceedings of the 42nd International Conference on Machine Learning, Vancouver, Canada
Identifiable Object Representations under Spatial Ambiguities
Jun 09, 2025Abstract:Modular object-centric representations are essential for *human-like reasoning* but are challenging to obtain under spatial ambiguities, *e.g. due to occlusions and view ambiguities*. However, addressing challenges presents both theoretical and practical difficulties. We introduce a novel multi-view probabilistic approach that aggregates view-specific slots to capture *invariant content* information while simultaneously learning disentangled global *viewpoint-level* information. Unlike prior single-view methods, our approach resolves spatial ambiguities, provides theoretical guarantees for identifiability, and requires *no viewpoint annotations*. Extensive experiments on standard benchmarks and novel complex datasets validate our method's robustness and scalability.
Average Calibration Losses for Reliable Uncertainty in Medical Image Segmentation
Jun 04, 2025Abstract:Deep neural networks for medical image segmentation are often overconfident, compromising both reliability and clinical utility. In this work, we propose differentiable formulations of marginal L1 Average Calibration Error (mL1-ACE) as an auxiliary loss that can be computed on a per-image basis. We compare both hard- and soft-binning approaches to directly improve pixel-wise calibration. Our experiments on four datasets (ACDC, AMOS, KiTS, BraTS) demonstrate that incorporating mL1-ACE significantly reduces calibration errors, particularly Average Calibration Error (ACE) and Maximum Calibration Error (MCE), while largely maintaining high Dice Similarity Coefficients (DSCs). We find that the soft-binned variant yields the greatest improvements in calibration, over the Dice plus cross-entropy loss baseline, but often compromises segmentation performance, with hard-binned mL1-ACE maintaining segmentation performance, albeit with weaker calibration improvement. To gain further insight into calibration performance and its variability across an imaging dataset, we introduce dataset reliability histograms, an aggregation of per-image reliability diagrams. The resulting analysis highlights improved alignment between predicted confidences and true accuracies. Overall, our approach not only enhances the trustworthiness of segmentation predictions but also shows potential for safer integration of deep learning methods into clinical workflows. We share our code here: https://github.com/cai4cai/Average-Calibration-Losses
Subgroups Matter for Robust Bias Mitigation
May 29, 2025Abstract:Despite the constant development of new bias mitigation methods for machine learning, no method consistently succeeds, and a fundamental question remains unanswered: when and why do bias mitigation techniques fail? In this paper, we hypothesise that a key factor may be the often-overlooked but crucial step shared by many bias mitigation methods: the definition of subgroups. To investigate this, we conduct a comprehensive evaluation of state-of-the-art bias mitigation methods across multiple vision and language classification tasks, systematically varying subgroup definitions, including coarse, fine-grained, intersectional, and noisy subgroups. Our results reveal that subgroup choice significantly impacts performance, with certain groupings paradoxically leading to worse outcomes than no mitigation at all. Our findings suggest that observing a disparity between a set of subgroups is not a sufficient reason to use those subgroups for mitigation. Through theoretical analysis, we explain these phenomena and uncover a counter-intuitive insight that, in some cases, improving fairness with respect to a particular set of subgroups is best achieved by using a different set of subgroups for mitigation. Our work highlights the importance of careful subgroup definition in bias mitigation and presents it as an alternative lever for improving the robustness and fairness of machine learning models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge